Ubigene’s gene KO E.coli model confirmed dietary D-xylose promotes intestinal health by phage activation

The elimination of intestinal pathogenic microorganisms is essential for the health of organisms. However, the complex intestinal community makes targeting specific bacterial organisms more challenging. There is increasing evidence that some foods can change the abundance of intestinal bacteria by regulating phages.
Professor Dandan Han’s research team from College of Animal Science and Technology, China Agricultural University, Beijing, China published an article entitled “Dietary D-xylose promotes intestinal health by inducing phage production in Escherichia coli” on Nature Journals npj Biofilms and Microbiomes, which used pathogenic Escherichia coli ATCC 25922 as a model to targeted knock out RecA and ldhA genes (constructed by Ubigene)[1]. It was found that dietary D-xylose could effectively reduce the survival rate of Escherichia coli ATCC 25922 in the intestine by activating prophage, providing a new idea for alleviating animal diarrhea and developing antibiotic substitutes.
First, three different prophages were identified in Escherichia coli ATCC 25922. The phage particles in the supernatant were examined by transmission electron microscopy. It was found that E. coli ATCC 25922 produced phages in LB medium containing D-xylose. Next, the effect of D-xylose on the phage production of E. coli ATCC 25922 was further evaluated. It was found that the addition of D-xylose could reduce the cell density (P < 0.05) (Fig. 1). With the increase of D-xylose concentration, the phage yield and phage: E. coli DNA ratio were also gradually increased, indicating that D-xylose could activate prophage in E. coli ATCC 25922 in vitro. The mice models infected by E. coli were supplemented with D-xylose, and it was found that it significantly reduced the number of E. coli ATCC 25922 in feces. These results showed that D-xylose promotes the production of phage in E. coli ATCC 25922 and inhibits the survival of E. coli ATCC 25922 in the gut.
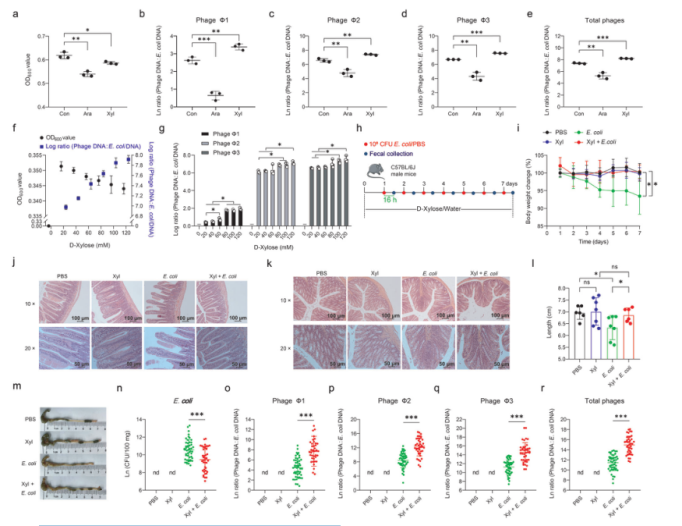
Figure 1. D-xylose promotes prophage induction in E. coli ATCC 25922
Next, the metabolic end products in the culture medium were analyzed. Results showed that D-xylose treatment significantly increased the production of D-lactic acid, formic acid and succinic acid, among which D-lactic acid was the most abundant end product (Fig. 2). In addition, with the increase of D-xylose concentration, the production of D-lactic acid also gradually increased, and the phage: Escherichia coli DNA ratio was significantly positively correlated with the level of D-lactic acid, indicating that D-lactic acid may be involved in the regulation of phage induction.
D-lactate dehydrogenase gene knockout E. coli (ΔLdhA mutant) constructed by Ubigene was used to find that the deletion of ldhA inhibited the production of D-lactic acid and reduced the content of phage. With the increase of D-lactic acid in LB medium, the cell density gradually decreased, indicating that D-xylose-mediated phage induction in E. coli ATCC 25922 was partially dependent on D-lactic acid. However, D-xylose in the diet decreased the lactic acid level and increased the propionic acid content in the feces of E. coli ATCC 25922-infected mice. Therefore, D-xylose drives different metabolic characteristics of lactic acid and propionic acid in vitro and in vivo.
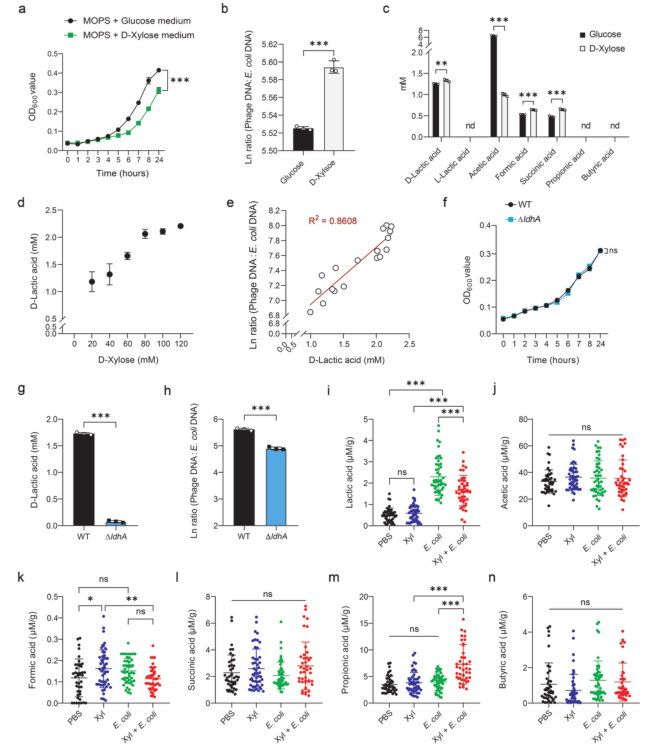
Figure 2. D-lactic acid correlates with prophage induction
Lactic acid is one of the substrates for propionic acid synthesis. The authors studied the sources of lactic acid that may be converted to propionic acid, and further explored the relationship between prophage induction and intestinal propionic acid levels. First, the mice were treated with oxamate, which blocked the production of L-lactic acid by inhibiting L-lactate dehydrogenase, and then infected with E. coli ATCC 25922 (Fig. 3). It was found that oxamate largely abolished the synthesis of L-lactic acid induced by E. coli, but had no effect on D-lactic acid. Then, the authors tested the effect of oxamate on D-lactic acid production of E. coli in vitro, and then supplemented the mice with D-xylose, and then treated them with oxamate and Escherichia coli. The results showed that it had no effect on D-lactic acid concentration, fecal propionic acid and phage: E. coli DNA ratio, indicating that L-lactic acid from the host is not a substrate for propionic acid synthesis or phage induction. After that, the authors infected mice with wild-type, ldhA-deficient and ldhA-overexpression mutant strains of ATCC 25922 and found that ldha+ mutant mice supplemented with D-xylose had the highest fecal propionic acid level, but the fecal propionic acid level of ΔldhA mutant mice decreased significantly, and the phage production also changed with the same trend as propionic acid. These results indicated that E. coli ATCC 25922 metabolizes D-xylose to produce D-lactic acid, which is in turn utilized by gut microbiota to produce propionic acid. The subsequent authors confirmed that Clostridia is responsible for the conversion of D-lactic acid to propionic acid in the intestine.
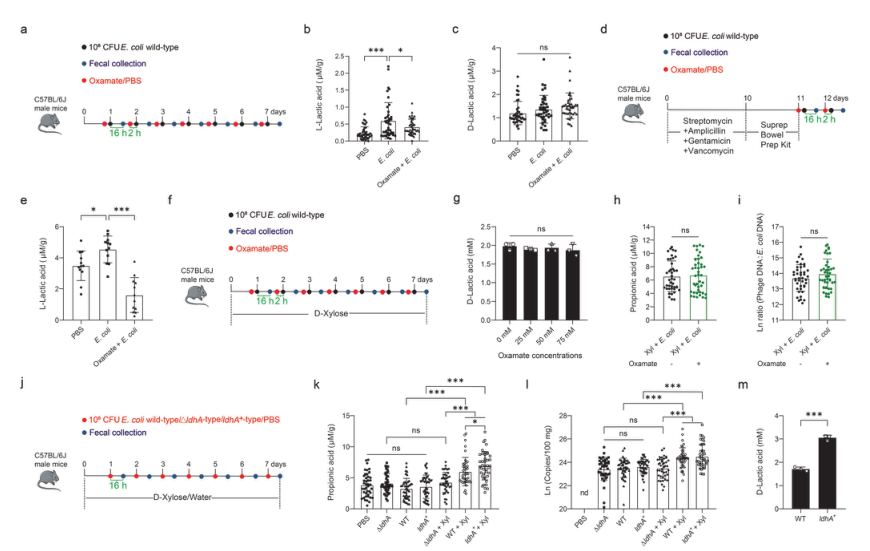
Figure 3. D-lactic acid metabolized by Escherichia coli ATCC 25922 is a substrate required for propionic acid synthesis
Later, the authors found that propionic acid could promote the induction of prophages in E. coli ATCC 25922 by adding propionic acid or sodium propionate to E. coli LB medium and dietary sodium propionate to the drinking water of E. coli infected mice.
Many phages benefit from the silencing of the SOS response to maintain a stable state. The activation of the SOS system is usually related to the production of phages. In E. coli, the key regulatory protein of the SOS system is RecA. To determine whether phage induction in E. coli ATCC 25922 is dependent on RecA, the authors used the ΔrecA knockout strain constructed by Ubigene, it was found that the phage production of the knockout strains was significantly reduced and grew better in MOPS medium with D-xylose as the sole carbon source or LB medium supplemented with sodium propionate (Fig. 4). This indicates that prophage induction in E. coli ATCC 25922 is dependent on RecA. Mice were gavaged with the wild-type or ΔrecA knockout strain and a more severe inflammation was observed in the ΔrecA knockout strain group, with higher levels of E. coli ATCC 25922 and lower levels of phage in feces. Taken together, these data suggest that dietary D-xylose or exposure to propionic acid triggers phage production by inducing the SOS response.
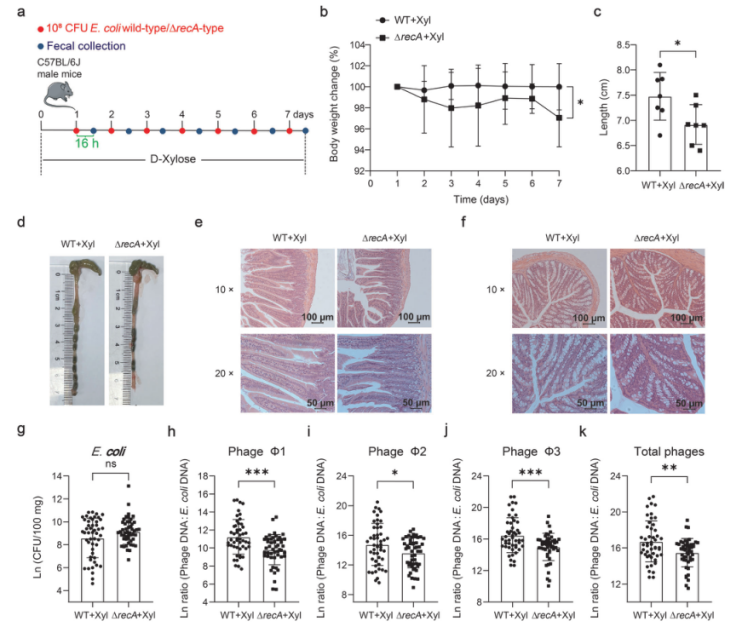
Figure 4. Inhibition of phage induction by Escherichia coli ATCC 25922 can worsen the intestinal inflammatory response
Few studies have reported the effects of dietary D-xylose on the composition of intestinal microorganisms and phage changes. The authors provided strong experimental evidence that dietary D-xylose reduces the survival rate of Escherichia coli ATCC 25922 by promoting the production of phages, thereby promoting intestinal health. The relevant mechanisms were elucidated, and the molecular framework of improving animal and human intestinal health by regulating temperate phages was revealed, At the same time, it provided a new strategy for developing dietary phage inducers as a treatment for pathogen infection or intestinal microbes.
Reference:
[1] Hu J, Wu Y, Kang L, et al. Dietary D-xylose promotes intestinal health by inducing phage production in Escherichia coli[J]. npj Biofilms and Microbiomes, 2023, 9(1): 79.










