CRISPR Library Screening: Are You Aware of These Details?


CRISPR Library Screening: Are You Aware of These Details?

As a powerful tool for screening functional genes, CRISPR library screening has demonstrated remarkable capabilities across various research fields. With this technology, researchers have successfully uncovered thousands of new targets, leading to hundreds of top-tier publications (CNS). Many scientists are also eager to use CRISPR screening to find their desired targets. However, undesirable results and difficulties in pinpointing targets often arise during the screening process, resulting in wasted time and funding. The details of the experiment are the determining factors for success and the reliability of the results. So, what aspects are crucial in CRISPR library screening?
This article outlines the key considerations for CRISPR library screening, helping you obtain better screening data!
1. Strict control of cell infection efficiency
The prerequisite for reliable screening results is that each cell introduces only a single gene mutation. Therefore, when constructing library cells, it is essential to ensure that each cell integrates only one sgRNA. Although a high viral infection efficiency can increase sgRNA coverage, it significantly raises the risk of multiple sgRNAs integrating into a single cell. Typically, it is recommended to control the viral infection efficiency at around 30%, which effectively avoids multiple infections while ensuring sgRNA coverage in the library cells.
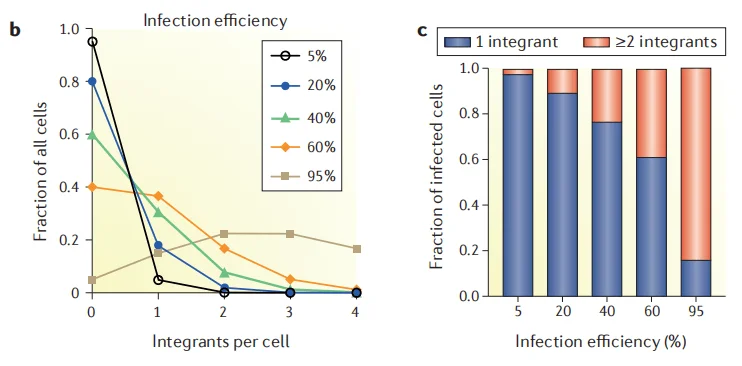
Figure 1: The higher the infection efficiency, the greater the probability of cells entering multiple sgRNAs. [1]
2. Ensuring sgRNA coverage in library cells
If the sgRNA coverage in library cells is inadequate before functional screening, potential target genes (corresponding sgRNAs) may be lost prior to screening, resulting in distorted results. Increasing the number of initial infected cells can effectively enhance sgRNA coverage and reduce random loss during passaging. Generally, it is recommended that the initial infected cell amount exceed 300 times the number of sgRNAs (300x coverage). The calculation formula for the required amount of wild-type cells is: Number of sgRNAs × 300 / infection efficiency (e.g., 0.3).
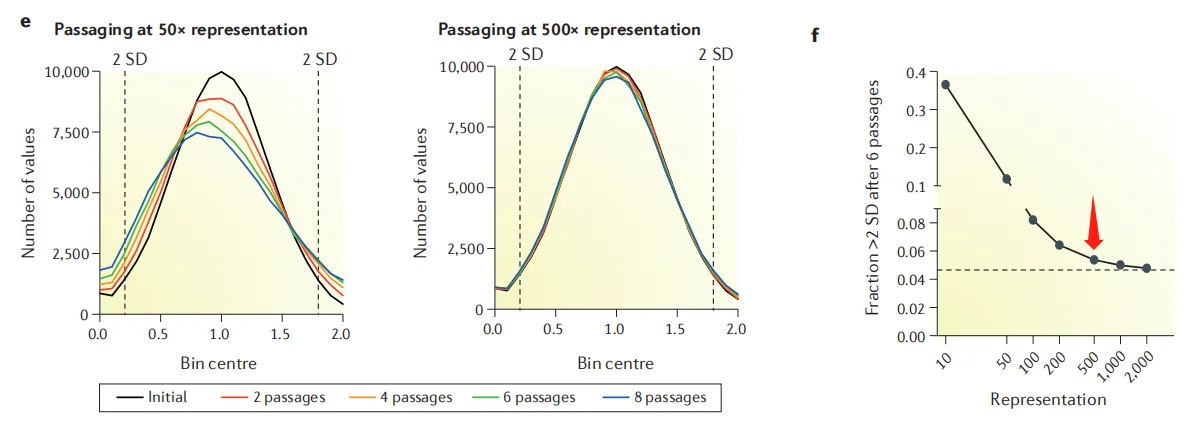
Figure 2: The more infected cells, the lower the proportion of sgRNA loss.
3. Increasing the initial amount of screening cells
Screening is essentially a process of enriching target cells. During the enrichment process, especially in projects with short screening cycles or low screening pressure, non-target cells are inevitably enriched alongside target cells, which dilutes the proportion of target cells and increases identification difficulty. Increasing the initial screening cell amount can significantly enhance the enrichment of target cells, reduce random error interference, and make the following bioinformatics analysis easier to identify actual target genes.
4. Increasing the number of sgRNAs targeting a single gene
The heterogeneity of CRISPR library cells depends on the editing efficiency of sgRNAs, but not all designed sgRNAs can effectively edit the target gene. In screenings, each effective sgRNA can be viewed as an independent replicate experiment. If the number of sgRNAs designed for a single gene is insufficient, the impact of “ineffective” sgRNAs on the final results will be magnified. To reduce interference from ineffective sgRNAs and enhance the credibility of results, it is recommended to design at least five sgRNAs for each gene.
5. The impact of cell gene editing efficiency
The editing efficiency of the CRISPR/Cas9 system is influenced by various factors, including sgRNA sequence, cell type, and culture time. If only 10% of the library cells are successfully edited, it means that 90% of the cells are functionally “wild-type”. These “wild-type” cells will neither be enriched nor lost in screenings, which will can severely dilute the enrichment signal of target cells or obscure the loss signal, creating significant interference for result analysis. Before screening, extending the culture time of library cells (1-2 weeks) can increase the proportion of edited cells, thereby reducing this factor's impact.
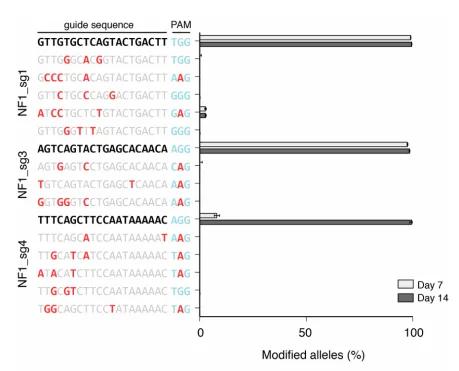
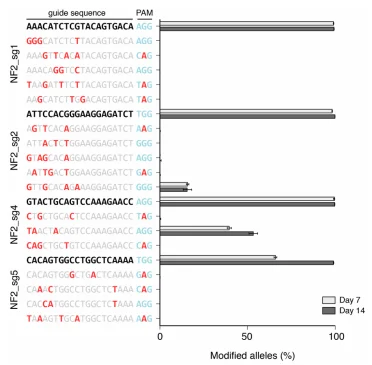
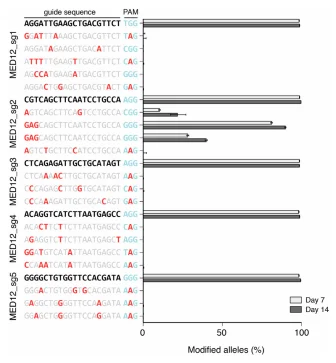
Figure 3: The effect of culture duration on gene editing efficiency.
6. The influence of screening intensity on the experiments.
CRISPR library screening can be divided into positive selection and negative selection:
- · Positive Selection: High screening pressure (e.g., drug treatment, nutrient deprivation) is applied, High screening pressure (e.g., drug treatment, nutrient deprivation) is applied, causing most cells to die; only a few cells with specific phenotypes (e.g., drug resistance) survive and are enriched. The sgRNA distribution in the enriched cell population is analyzed to identify target genes (e.g., resistance genes).
- · Negative Selection: Weaker screening pressure (e.g., low-dose drugs) is applied, leading to only a small number of pressure-sensitive cells dying while most cells survive. The lost sgRNA distribution is analyzed to identify target genes (e.g., drug-sensitive genes).
Screening intensity directly impacts the characteristics of results and analysis method:
| IntensityScreening Type | Screening | Characteristics/th> | Bioinformatics Analysis Method | Target Gene sgRNA Variation Features (Experimental Group vs Control Group) | Target Gene | Result Credibilit |
|---|---|---|---|---|---|---|
| Positive Selection | High | Most cells are lost during screening; collect remaining enriched cells for NGS sequencingNGS | Positive Screening | Significant enrichment | Resistance Genes | High |
| Negative Screening | Significant loss | Drug-sensitive Genes | Low | |||
| Negative Selection | Low | A few cells are lost during screening; collect a large number of surviving cells for NGS sequencing | Positive Screening | Enrichment | Resistance Genes | Low |
| Negative Screening | Significant loss | Drug-sensitive Genes | Higher |
Key Interpretations:
- · In positive Selection: positive analysis (seeking enriched sgRNAs) focuses on surviving cells. The number of enriched sgRNAs is small, the target gene signal is concentrated and significant (e.g., resistance genes), making the results highly credible.
- · Innegative selection, negative analysis (looking for lost sgRNAs) targets dead cells. The number of differential genes is relatively low, and the signals are clear (e.g., drug-sensitive genes), hence results are relatively credible.
- · Conversely, negative analysis in positive selection (analyzing lost sgRNAs) yields a large number of differential genes with weak signals, whereas positive analysis in negative selection (analyzing enriched sgRNAs) often reflects differences in cell proliferation or random errors rather than true resistance mechanisms. Both scenarios yield results with lower credibility.
7. Acknowledging the reliability differences between positive and negative selection results
As described, there are inevitably some “wild-type” cells in the library cell pool that carry sgRNAs but are not successfully edited. These cells affects the results differently for each type of screening:
- · In negative selection, we focus on lost cells. However, among the cells carrying target gene sgRNAs, only the successfully edited sensitive cells will be lost, while unedited “wild-type” cells remain alive. This seriously dilutes the loss signal from target gene sgRNAs, reducing the significance of differential analysis and complicating target gene identification.
- · In positive selection, we focus on surviving/enriched cells. Under strong screening pressure, only the successfully edited target cells (e.g., those gaining resistance) will survive and be enriched (usually undergoing rounds of screening and proliferation), while both unedited “wild-type” cells and non-target cells are eliminated. Therefore, the “wild-type” cells has a minimal impact on the positive analysis results of positive selection.
Thus, the positive analysis results from positive selection in CRISPR library screening are usually more credible than the negative analysis results from negative selection, especially after multiple rounds of enrichment
Summary
The reliability of CRISPR library screening results is affected by multiple factors, including cell gene editing efficiency, sgRNA quantity and design, screening pressure intensity, and data analysis strategies. Understanding and meticulously controlling these critical details is fundamental to obtaining successful and reliable screening data.
Ubigene has a professional CRISPR library screening team that can help you achieve more reliable screening results! Feel free to inquire>>>
References
[1] Doench JG. Am I ready for CRISPR? A user's guide to genetic screens. Nat Rev Genet. 2018 Feb;19(2):67-80.
[2]Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014 Jan 3;343(6166):84-87.


