CRISPR Screening | A Comprehensive Guide to Building a Functional Screening System


CRISPR Screening | A Comprehensive Guide to Building a Functional Screening System

CRISPR screening is a revolutionary technology that enables high-throughput, precise, and systematic identification of genes associated with specific biological phenotypes or functions on a large scale, including genome-wide analysis. This approach represents a shift from broad, undirected gene perturbation toward the precise dissection of gene functions and phenotypes. In this context, establishing a robust and tailored functional screening system that accurately models the research context becomes critically important.
Functional Screening: A Selective Pressure-Based Assay Mimicking Cellular Darwinism
In essence, functional screening involves applying a defined selective pressure to a pre-constructed cell pool, thereby inducing the emergence of a desired phenotype—such as drug resistance, migration, activation, or cell death. Through carefully designed selection strategies, cell populations exhibiting the target phenotype are efficiently enriched.
Key Features of CRISPR-U™:
Under selective pressure, cells that gain a functional advantage—such as increased drug resistance or enhanced proliferation—through genetic perturbations (e.g., gene knockout, activation, or inhibition) will preferentially proliferate Conversely, disadvantaged cells—those more sensitive or growth-arrested—will be outcompeted or eliminated. This process closely mimics the principles of natural selection. By sequencing and analyzing the sgRNAs enriched in the surviving population, researchers can identify precisely key genetic drivers underlying the selected phenotype.
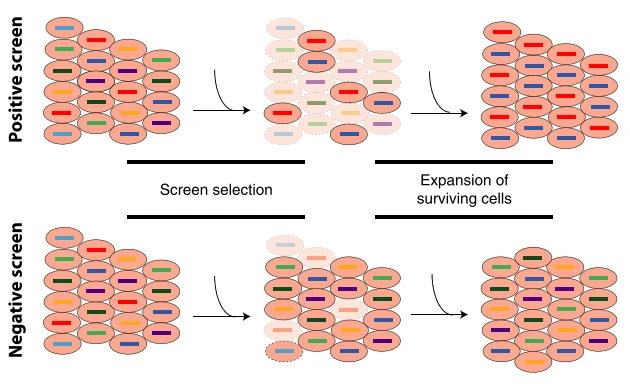
Figure 1. Core Mechanism of Functional Screening — "Survival of the Fittest" [1]
Establishing a Functional Screening System
Functional screening system is tailored to specific research objectives and the phenotype of interest. A well-designed screening platform consists of two major components: selective pressure application and phenotype enrichment.
Selective Pressure — Simulating a "Physiological Challenge"
Selective Pressure Application refers to the strategic design and implementation of precise screening conditions tailored to the specific research objectives, which aims to induce the desired phenotypic outcome in the cell population. Ubigene offers a diverse range of customizable selective pressure options to effectively model relevant biological contexts.
- 1. Serial Passaging — "Natural Evolution Under Basal Conditions": Through extended culture and serial passaging, cells harboring adaptive genetic variants gain a proliferative or survival advantage and become progressively enriched within the population. Conversely, cells carrying sensitivity- or lethality-associated mutations exhibit impaired growth or viability, leading to their gradual depletion or elimination.
- 2. Drug Treatment — "Pharmacological Selection": By exposing cells to specific drug compounds, selective pressure is applied to induce corresponding phenotypic responses. Cells harboring drug resistance-associated genetic variants gain a survival and proliferative advantage, leading to their progressive enrichment. In contrast, cells with sensitivity- or lethality-related mutations exhibit impaired growth or viability, resulting in their gradual depletion or elimination.
- 3. Viral Infection — “Host-Pathogen Interaction”: 3.By exposing cells to specific pathogens or viruses, selective pressure is applied to elicit corresponding phenotypic responses. Cells harboring adaptive genetic variants that confer resistance or survival advantages become progressively enriched, while cells carrying sensitivity- or lethality-associated mutations experience impaired growth or viability and are gradually depleted or eliminated.
- 4. Functional Cell Co-culture — “Cellular Competition”: This approach involves introducing specific functional cell types—such as effector immune cells—into the culture system alongside target cells, for example, co-culturing immune effector cells with tumor cells. Through this co-culture interaction, selective pressure is applied that induces the desired phenotypic response in the target cells. Cells harboring adaptive genetic variants gain a growth or survival advantage and become progressively enriched, whereas cells with sensitivity- or lethality-associated mutations exhibit reduced fitness, leading to their gradual depletion or elimination.
- 5. Cytokine Stimulation — “Signaling Storm”: By treating cells with specific growth factors or small-molecule compounds, selective pressure is applied to induce corresponding phenotypic changes. Cells harboring adaptive genetic variants gain a proliferative or survival advantage and become progressively enriched, whereas cells carrying sensitivity- or lethality-associated mutations exhibit impaired growth or viability, leading to their gradual depletion or elimination.
- 6. Other Treatment Modalities — “Extreme Challenge”: Additional selective pressures, such as hypoxia or nutrient deprivation, can be applied to mimic harsh microenvironmental conditions. For customized screening system design tailored to specific research needs, please contact Ubigene for technical support. We offer personalized solutions to develop and implement functional screening platforms aligned with your experimental objectives.
Phenotypic Enrichment — Precise “Selection of targets”
Phenotypic enrichment refers to the process of isolating and capturing cells exhibiting the desired phenotype induced by the applied selective pressure. This step is critical for refining the cell population to those most relevant to the screening objective. Ubigene offers a range of state-of-the-art enrichment methodologies widely adopted in the field:
- 1. Harvesting Cells After Defined Culture Period (Based on Proliferation/Apoptosis): 1.Following the application of selective pressure, cells are cultured for a specified duration to allow phenotypic manifestations such as altered proliferation or survival. At the end of this period, the cell population is harvested, and changes in sgRNA representation are assessed based on cell abundance. This approach enables evaluation of sgRNA enrichment or depletion correlated with differential cell fitness under the given conditions.
- 2. Cell Harvesting via Flow Cytometry or Magnetic Sorting (Based on Expression Levels of Cell-Specific Markers): Following selective pressure, cells exhibiting differential expression of specific markers are isolated using flow cytometry (FACS) or magnetic-activated cell sorting (MACS). By segregating populations with high versus low marker expression, sgRNA enrichment and depletion can be evaluated in correlation with phenotypic marker status, enabling precise identification of genetic perturbations driving the observed phenotype.
- 3. Cell Harvesting Based on Specific Substrate-Mediated Separation (Behavioral Phenotypes): Following selective pressure, cells exhibiting distinct behavioral phenotypes—such as migration capacity or adhesion properties—are isolated using substrate- or matrix-based separation techniques. This approach enables the enrichment of target cell populations based on functional behavior, allowing subsequent evaluation of sgRNA enrichment or depletion associated with these phenotypic traits.
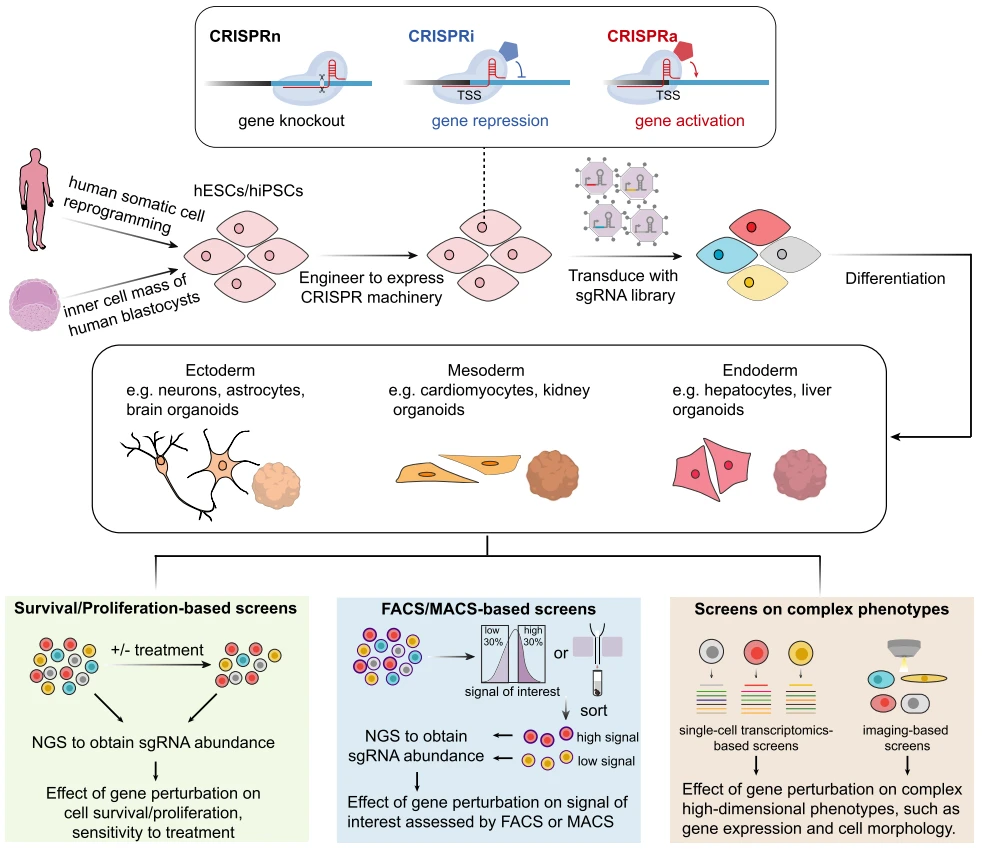
Figure 2. CRISPR Screening Workflow and Functional Screening Platform [2]
Having clarified the core mechanisms and construction strategies of functional screening, researchers may have already formed a preliminary framework for their studies. However, despite its powerful capabilities, functional screening also presents significant challenges, including large experimental scale, difficulty in optimizing conditions, and high technical barriers.
Key Challenges in Functional Screening:
- 1. Large-scale cell culture requirements, leading to substantial demands for cultivation equipment and consumables;
- 2. Extensive preliminary experiments needed to optimize screening conditions;
- 3. High technical demands on cell culture techniques and environmental conditions;
- 4. Reliance on specialized expertise and operational experience related to functional screening platforms;
- 5. Necessity for meticulous experimental workflow design and process control;
- 6. Careful integration and coordination of all stages within the CRISPR screening pipeline.
Ubigene's Diversified Phenotypic Screening & Analysis Platform
By orthogonally combining various types of selective pressures and enrichment strategies, a wide array of functional screening systems can be derived. Researchers can customize their screening platforms by selecting the most appropriate “selective pressure application” and “phenotypic enrichment” methods tailored to their specific scientific objectives. To address diverse research needs, Ubigene has established a comprehensivephenotypic screening and analysis platform,offering refined and specialized functional screening services to support our clients' projects with high precision and flexibility.
Diversified Phenotypic Analysis Platform
Ubigene’s diversified phenotypic analysis platform supports a broad range of functional screening systems, comprehensively covering key screening operations such as drug and viral treatments, serial passaging, co-culture assays, flow cytometric sorting, cell migration, and adhesion-based selections.
Robust and Mature Cell Biology Platform
Ubigene’s cell biology platform is equipped to meet the demands of large-scale cell culture, staffed by skilled technicians with extensive cell culture expertise. This platform supports functional screening across a variety of cellular models, ensuring reliability and reproducibility in experimental outcomes.
Comprehensive End-to-End Service
Ubigene offers a well-established system for preliminary optimization and quality management, delivering full-process functional screening and one-stop CRISPR screening services. Our integrated workflow ensures seamless coordination across all stages of the CRISPR screening pipeline, maximizing experimental continuity and success rates.
Expert Technical and Project Management Support
Ubigene is backed by a team of PhD-level specialists with extensive expertise and hands-on experience in functional screening platforms. Our experts provide personalized, step-by-step guidance throughout the design and execution of functional screening projects to ensure optimal results.
Workflow:
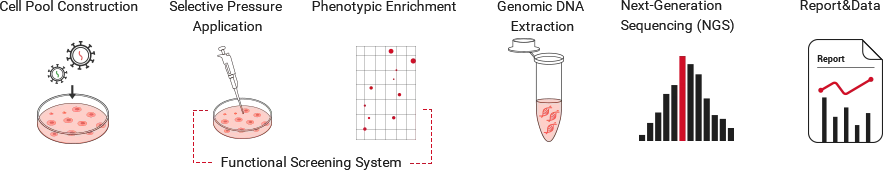
Application Scenarios (Examples):
1. Serial Passaging + Enrichment Based on Cell Proliferation/Apoptosis
This screening system is commonly applied for:
- A. Identification of genes essential for cell growth and survival;
- B. Screening of synthetic lethal targets.
2. Serial Passaging + Enrichment Based on Cell-Specific Marker Expression Levels
This screening system is commonly applied for:
- A. Antigen epitope identification;
- B. Investigation of signaling pathway regulatory mechanisms;
- C. Studies on single-gene regulatory mechanisms.
3. Serial Passaging + Enrichment Based on Behavioral Phenotypes
This screening system is typically applied to uncover novel regulators associated with specific cellular behaviors, such as:
- A. Regulators of cell migration;
- B. Regulators of cell adhesion;
4. Drug Treatment + Specific Enrichment Method
This screening system is commonly applied for:
- A. Identification of tumor drug resistance genes;
- B. Screening of genes involved in drug synergy;
- C. Investigation of drug targets and mechanisms of action.
5. Viral Infection + Specific Enrichment Method
This screening system is commonly applied for:
- A. Identification of host factors involved in viral infection;
- B. Screening of genes that synergize with vaccine responses;
- C. Investigation of vaccine targets and mechanisms of action.
6. Functional Cell Co-culture + Specific Enrichment Method
This screening system is commonly applied for:
- A. Identify genes regulating immune cell persistence and resistance to exhaustion;
- B. Identify genes controlling tumor cell sensitivity to immune-mediated cytotoxicity;
- C. Identify genes involved in immune cell activation-induced cell death;
- D. Identify genes influencing survival advantages related to cytokine or nutrient competition.
7. Cytokine Stimulation + Specific Enrichment Method
This screening system is commonly applied for:
- A. Identify factors that promote or inhibit specific cellular biological processes;
- B. Investigate regulatory mechanisms of signaling pathways;
- C. Study targets and mechanisms of cytokine action.
Although in vitro functional screening systems are currently more widely used due to their higher stability and flexibility—and are highly efficient for preliminary screening, mechanistic studies, and large-scale target discovery—in vivo screening is indispensable for translational research, critical target validation, and evaluation of complex phenotypes such as immunity, metastasis, drug resistance, and systemic effects. The core advantage of in vivo functional screening lies in its unparalleled physiological and pathological relevance. It reveals gene functions within the context of the whole organism, complex microenvironments, and dynamic systems. This is especially crucial for studying immune interactions, tumor metastasis, pharmacokinetics/pharmacodynamics (PK/PD), systemic effects, and host factors—areas where in vitro screening falls short. Compared to in vitro screening, in vivo screening offers the following key benefits:
- 1. Recapitulates authentic physiological and pathological environments;
- 2. Integrates the role of the immune system;
- 3. Provides more accurate insights into drug responses and resistance;
- 4. Evaluates in vivo adaptation and selective pressures;
- 5. Identifies genes associated with metastasis and colonization;
- 6. Assesses therapeutic windows and potential toxicity in live organisms;
- 7. Enables identification of host factors relevant in vivo.
Challenges to Consider:
Certainly, in vivo screening also presents several challenges: higher costs, longer experimental timelines, increased technical complexity (e.g., efficient sgRNA library delivery, adherence to animal ethics regulations), and more complicated data analysis due to background noise and biological heterogeneity. Additionally, throughput is generally lower compared to in vitro screening. Therefore, establishing an in vivo screening system requires more meticulous and comprehensive experimental design to ensure the stability and accuracy of the screening platform. For more information on diverse functional screening applications, please feel free to contact Ubigene. We are committed to providing you with detailed materials and expert support.

Figure 4. In Vitro and In Vivo Functional Screening Platforms [3]
Application
Functional Screening System: Serial Passaging (Selective Pressure) + Cell Survival/Proliferation (Enrichment Method)
Application: Identification of essential genes for the growth of human melanoma cells and pluripotent stem cells
To date, this type of functional screening system has become less common, primarily because it was extensively utilized during the early development of CRISPR screening platforms to identify essential genes required for cell growth or proliferation across various cell types. In this study, the authors successfully employed this screening approach to pinpoint genes essential for the proliferation of human melanoma cells and human pluripotent stem cells. This work has significantly advanced CRISPR screening technology and provided valuable insights and foundational knowledge for the field of tumor biology.
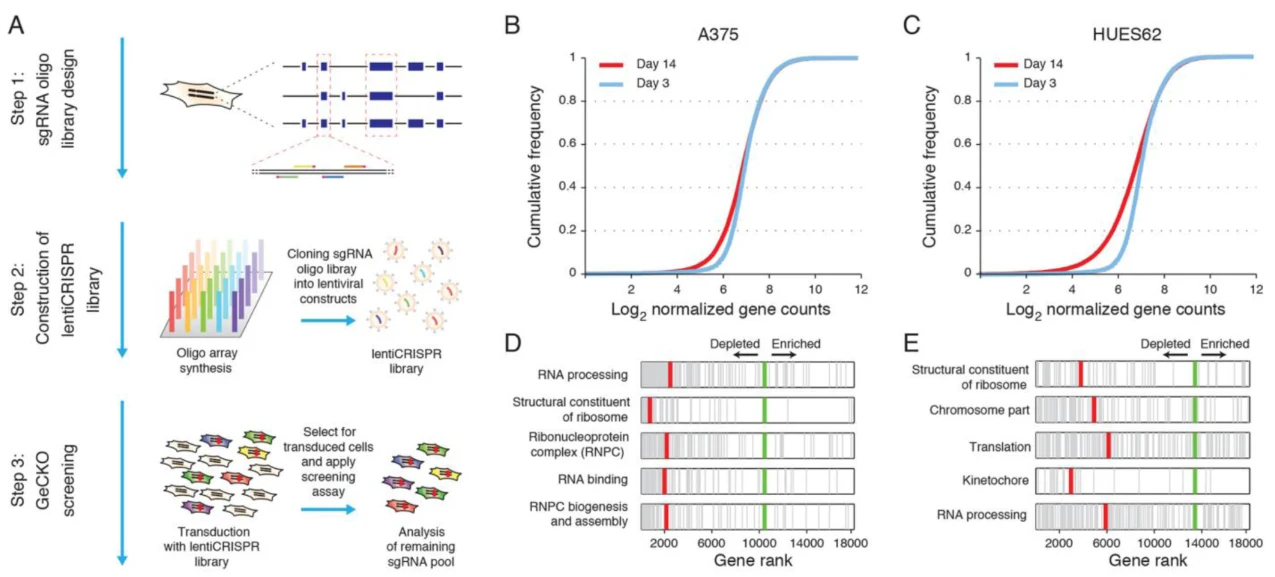
Figure 1. Functional Screening System for Identifying Genes Essential for Cell Growth and Proliferation
Functional Screening System: Serial Passaging + Enrichment Based on Cell-Specific Marker Expression Levels
Application: Investigation of Single-Gene Regulatory Mechanisms
In this study, the authors established a CRISPR screening platform targeting 96 members of the deubiquitinase enzyme family. By analyzing cell populations with low and high PD-L1 expression levels, they identified ATXN3 as a positive transcriptional regulator of PD-L1. Tumors lacking ATXN3 showed improved responses to low-dose anti-PD-1 therapy, and its inhibition enhanced the efficacy of immune checkpoint blockade treatment.
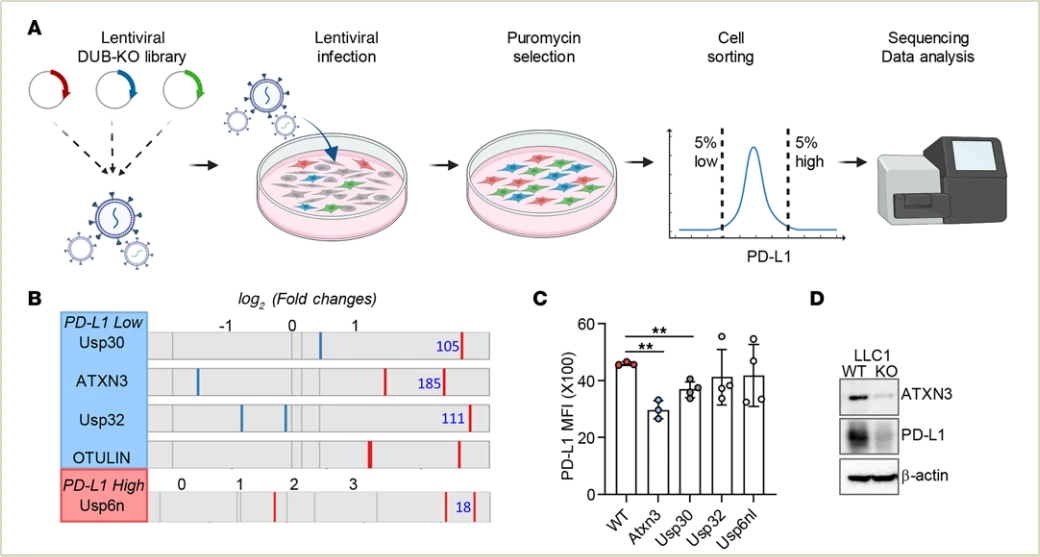
Figure 2. Functional Screening System for Identifying Upstream Regulatory Targets of Specific Genes
Functional Screening System: Drug Treatment (Selective Pressure) + Cell Survival/Proliferation (Enrichment Method)
Application: Mechanistic Investigation of the Anti-Glioma Compound Gliocidin
In this study, the authors utilized the mouse whole-genome CRISPR knockout library (Brie) to perform a functional screen in NG2-3112 glioma cells. Cells were treated with different concentrations of Gliocidin (IC50 and IC80), and samples were collected on Day 0 and Day 14 post-treatment. Both positive and negative selection analyses revealed that perturbation of mTORC1 pathway components significantly altered cell sensitivity to Gliocidin: positive regulators of mTORC1 reduced sensitivity, while negative regulators enhanced it. These findings highlight the critical role of the mTORC1 pathway in mediating the anti-tumor efficacy of Gliocidin in brain cancer.
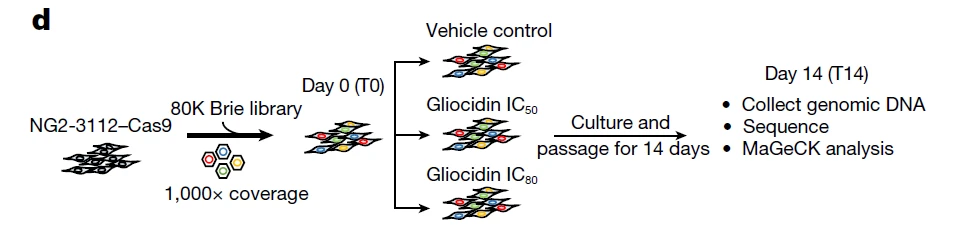
Figure 3. Functional Screening System for Identifying Drug Mechanisms of Action and Targets
Functional Screening System: Drug Treatment (Selective Pressure) + Flow Cytometric Sorting (Enrichment Method)
Application: Mechanistic Study of Ternatin-4–Mediated Degradation of eEF1A
In this study, the authors engineered a cell line overexpressing an mCherry-eEF1A fusion protein and conducted a CRISPRi screen targeting the ubiquitin-proteasome system (UPS) gene set. Building on prior evidence that Ternatin-4 promotes eEF1A degradation, the authors treated the cells with Ternatin-4 and sorted the populations based on mCherry fluorescence intensity to distinguish between cells with high and low levels of eEF1A. This fluorescence-based enrichment strategy enabled the identification of key regulators—specifically RNF14 and RNF25—as critical E3 ligases involved in the ubiquitination of eEF1A and ribosomal proteins, shedding light on the molecular mechanism by which Ternatin-4 exerts its activity.
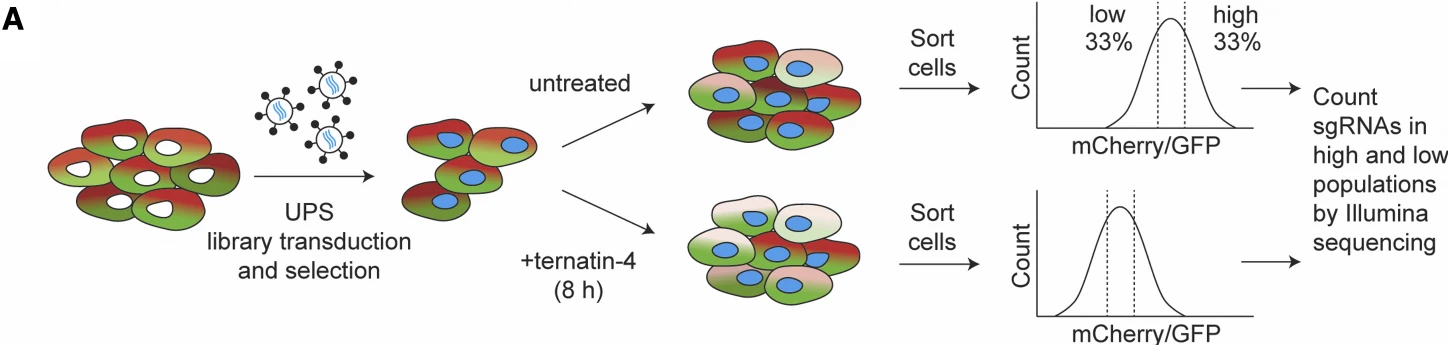
Figure 4. Dissecting Signaling Mechanisms Using Drug Treatment Combined with Flow Cytometric Sorting
Functional Screening System: Drug Treatment (Selective Pressure) + Behavioral Phenotype (Enrichment Method)
Application: Identification of Genes Involved in Adhesion-Dependent and -Independent Cell Migration, Protein Transport, and Actin Cytoskeleton Regulation
In this study, the authors employed a genome-wide CRISPRi library and designed three distinct experimental models to investigate key regulators of various modes of neutrophil migration. In the first two models, cells were seeded into the upper reservoir of transwell chambers equipped with track-etched membranes containing 3 μm pores. By applying 10% heat-inactivated fetal bovine serum (FBS) at specific reservoir locations, the authors established chemotactic gradients to evaluate directed (oriented) and random (non-oriented) migration behaviors. In the third model, cells were embedded in synthetic extracellular matrix to simulate amoeboid-like 3D migration, mimicking how cells traverse interstitial tissue spaces. Through these models, the screen identified 344 gene knockdowns that inhibited migration and 31 that enhanced it. Importantly, the study uncovered a critical role for mTORC1 signaling in HL60 cell differentiation, demonstrating its influence on neutrophil abundance, viability, and migratory behavior.
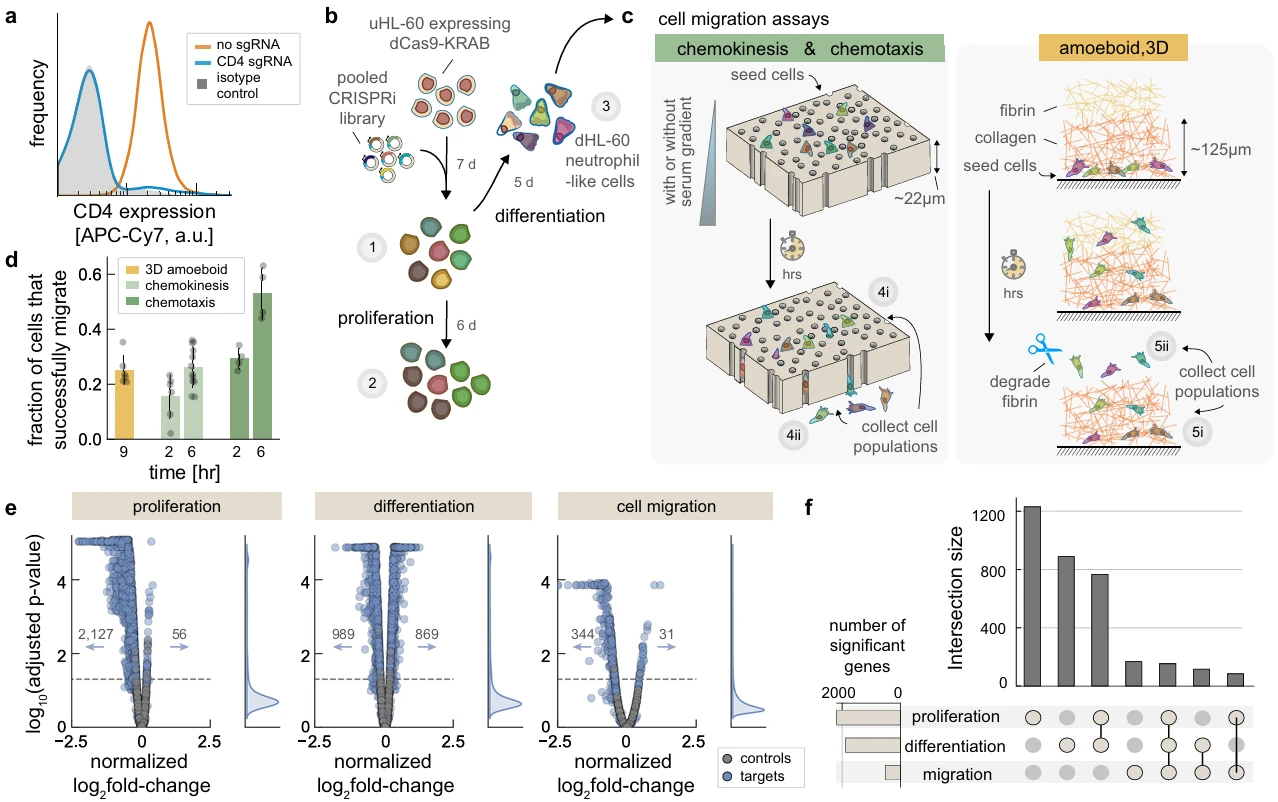
Figure 5. Functional Screening Models for Studying Cell Migration
Functional Screening System: Viral Infection (Selective Pressure) + Cell Survival/Proliferation (Enrichment Method)
Application: Identification of Host Antiviral Targets
In this study, the authors engineered more than 1,500 replication-competent HIV-1 viruses, each carrying a unique sgRNA targeting over 500 host genes, to perform a CRISPR-based functional screen. The screen aimed to identify sgRNAs that enhance HIV-1 replication fitness. By passaging these viruses in Cas9-expressing CD4⁺ T cells and performing next-generation sequencing, the researchers identified several host restriction factors—including GRN, CIITA, and EHMT2—that significantly suppress HIV-1 replication. These findings provide important insights into host-virus interactions and uncover potential antiviral therapeutic targets.
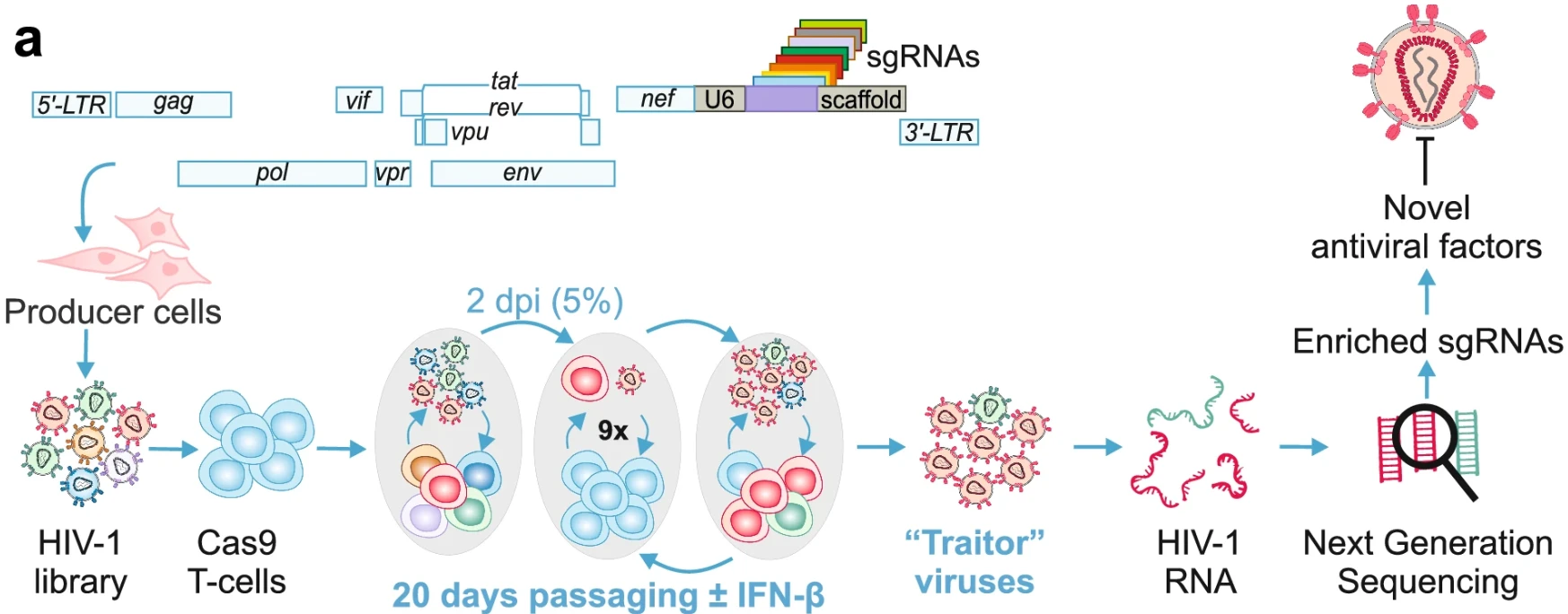
Figure 6. Functional Screening Model for Identifying Antiviral Host Factors
Functional Screening System: Tumor/Immune Cell Co-culture (Type of Pressure) + Cell Survival/Proliferation (Enrichment Strategy)
Application: Identification of Immune Resistance Regulators
In this study, the authors employed a co-culture system of murine colon cancer cells (MC38) and PmelT T cells as the applied selective pressure, aiming to identify novel genes that critically regulate therapeutic efficacy in immunotherapy. During the functional screen, PmelT cells were added to the tumor cells at effector-to-target (E:T) ratios of 0.3:1 and 1:1 and co-cultured for 16 hours to induce immune-mediated cytotoxic pressure. For the non-T cell control group, an equivalent volume of T cell culture medium was added to assess baseline sensitivity or resistance to T cell–mediated cytotoxicity in vitro. The study revealed two distinct immune resistance regulators, highlighting their potential as therapeutic targets to enhance immunotherapy efficacy. Among these, PRMT1 was identified as a dual immune resistance regulator, while RIPK1 functioned as a cytotoxic resistance regulator. Despite differential impacts across various immunotherapy modalities, targeting PRMT1 and RIPK1 genetically sensitized tumors to T cell–mediated killing and improved responses to anti–PD-1 and OX40 checkpoint blockade therapies.

Figure 7. Tumor/Immune Cell Co-culture System Reveals Key Regulators of Immunotherapy
Functional Screening System: Cytokine Stimulation (Pressure Type) + Cell Survival/Proliferation (Enrichment Method)&Cytokine Stimulation (Pressure Type) + Flow Cytometry Sorting (Enrichment Method)
Application: Identification of Key Regulatory Genes Associated with Activation of Aged Neural Stem Cells In Vitro
In this study, the authors performed a genome-wide CRISPR knockout screen on primary neural stem cells (NSCs) isolated from young and aged mice. Following lentiviral library transduction of quiescent NSCs, cytokines were applied to induce the transition of NSCs from a quiescent to an activated state, thereby enhancing their activation and proliferative capacity. For enrichment, two approaches were employed: the first involved flow cytometry sorting of Ki67-positive cells on day 4 post-activation to quantify sgRNA representation; the second was based on the proliferative advantage of activated NSCs, collecting cells on day 14 post-activation to analyze sgRNA abundance. Utilizing these complementary functional screening strategies, the authors successfully identified 301 key genes whose knockout specifically promotes activation of aged NSCs in vitro.
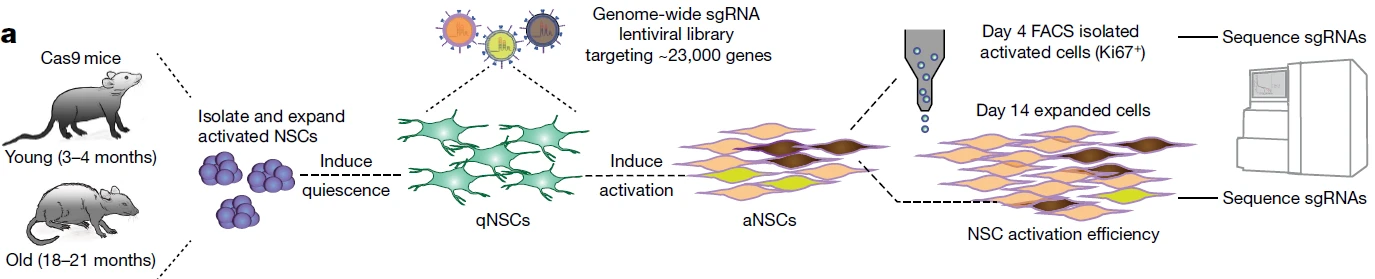
Figure 8. Dual-Functional Screening System Combined to Identify Key Regulators of Aged Neural Stem Cell Activation
Functional Screening System: Tumor/Immune Cell Co-culture (Type of Pressure) + Cell Survival/Proliferation (Enrichment Strategy)
Application: Identification of Immune Resistance Regulators
In this study, the authors employed a co-culture system of murine colon cancer cells (MC38) and PmelT T cells as the applied selective pressure, aiming to identify novel genes that critically regulate therapeutic efficacy in immunotherapy. During the functional screen, PmelT cells were added to the tumor cells at effector-to-target (E:T) ratios of 0.3:1 and 1:1 and co-cultured for 16 hours to induce immune-mediated cytotoxic pressure. For the non-T cell control group, an equivalent volume of T cell culture medium was added to assess baseline sensitivity or resistance to T cell–mediated cytotoxicity in vitro. The study revealed two distinct immune resistance regulators, highlighting their potential as therapeutic targets to enhance immunotherapy efficacy. Among these, PRMT1 was identified as a dual immune resistance regulator, while RIPK1 functioned as a cytotoxic resistance regulator. Despite differential impacts across various immunotherapy modalities, targeting PRMT1 and RIPK1 genetically sensitized tumors to T cell–mediated killing and improved responses to anti–PD-1 and OX40 checkpoint blockade therapies.
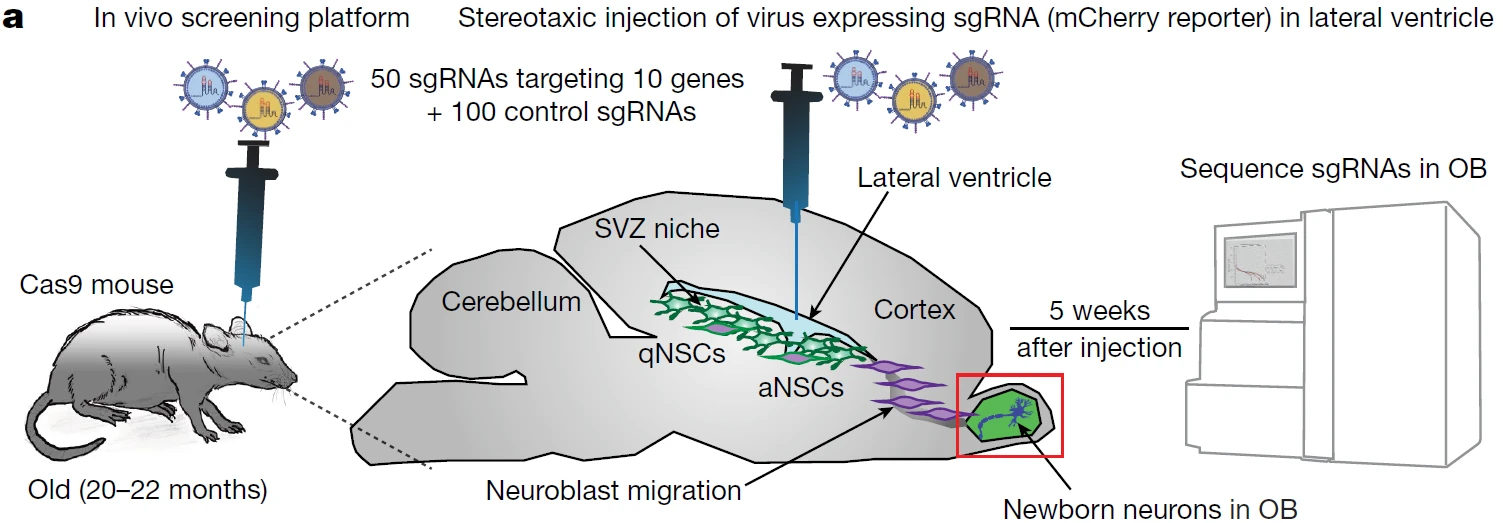
Figure 9. Efficient Validation of Key Regulators of Aged Neural Stem Cell Activation Using In Vivo Functional Screening
CRISPR Screen, flexible and on-demand — Ubigene is your trusted partner!
Whether you’re uncovering unknown mechanisms, identifying drug targets, tackling resistance challenges, or optimizing immunotherapy strategies, Ubigene’s functional screening technology platform and expert service team are your trusted partners. We are dedicated to providing you with:
- Personalized screening design: Tailored strategies (selective pressure application + phenotypic enrichment) customized to your specific scientific questions.
- Efficient and stable screening execution: Leveraging mature platforms and refined workflows to ensure high-quality data.
- Seamless integration of the full screening workflow: 3.Smooth connection of all CRISPR screening steps, guaranteeing stability and accuracy.
- Professional data analysis support: Empowering you to extract meaningful biological insights from massive datasets.
Reference
[1]Miles LA, Garippa RJ, Poirier JT. Design, execution, and analysis of pooled in vitro CRISPR/Cas9 screens. FEBS J. 2016 Sep;283(17):3170-80.
[2]Li K, Ouyang M, Zhan J, Tian R. CRISPR-based functional genomics screening in human-pluripotent-stem-cell-derived cell types. Cell Genom. 2023 Apr 18;3(5):100300.
[3]Chow RD, Chen S. Cancer CRISPR Screens In Vivo. Trends Cancer. 2018 May;4(5):349-358. doi: 10.1016/j.trecan.2018.03.002.


