A Practical Guide to Constructing Stable Knockout Cell Lines Using CRISPR/Cas9


A Practical Guide to Constructing Stable Knockout Cell Lines Using CRISPR/Cas9 — From sgRNA Design to Single- Cell Clone Validation
Introduction
CRISPR/Cas9 gene editing technology has become a core tool for investigating gene function, disease mechanisms, and drug target validation due to its high efficiency, simplicity, and programmable nature. Compared with transient gene editing, stable knockout cell lines can maintain consistent gene deletion across multiple passages, ensuring experimental reproducibility and reliability. This makes them indispensable models in both basic biological research and drug development. However, constructing a stable knockout cell line is a multistep and technically demanding process, involving several critical phases — from rational sgRNA design to precise single-cell clone selection and validation. This guide provides a systematic overview of the standard workflow for generating stable knockout cell lines using CRISPR/Cas9 and highlights how leveraging Ubigene’s specialized gene editing platform can help researchers achieve higher efficiency and precision in genome editing.
Pre-Experimental Preparation
Before initiating the experiment, it is essential to conduct a comprehensive assessment of the target gene and cell line characteristics to ensure that the experimental design is both scientifically sound and technically feasible. The key preparatory steps include:
- · Gene Information Confirmation:Examine the target gene for transcript variants, alternative splicing events, and functional domains. Public databases such as Ensembl and NCBI can be used to obtain detailed genomic and transcriptomic information.
- · Cell Line Characterization:Evaluate relevant cellular properties, including transfection efficiency, growth rate, drug sensitivity, and whether the cell type is considered “difficult-to-transfect” (e.g., primary cells, stem cells, immune cells, etc.).
- · Gene Editability Assessment:Determine whether the target gene is essential for cell viability or proliferation. This can be done by consulting public resources such as DepMap or gene essentiality databases, or by using Ubigene’s Gene Dependency Assessment System, to avoid lethality or severe growth defects resulting from gene knockout.
- · Downstream Validation Planning:Develop a detailed validation strategy prior to editing, including protein expression analysis (Western blot), quantitative PCR, functional assays, and Sanger sequencing to confirm the knockout efficiency and functional impact.
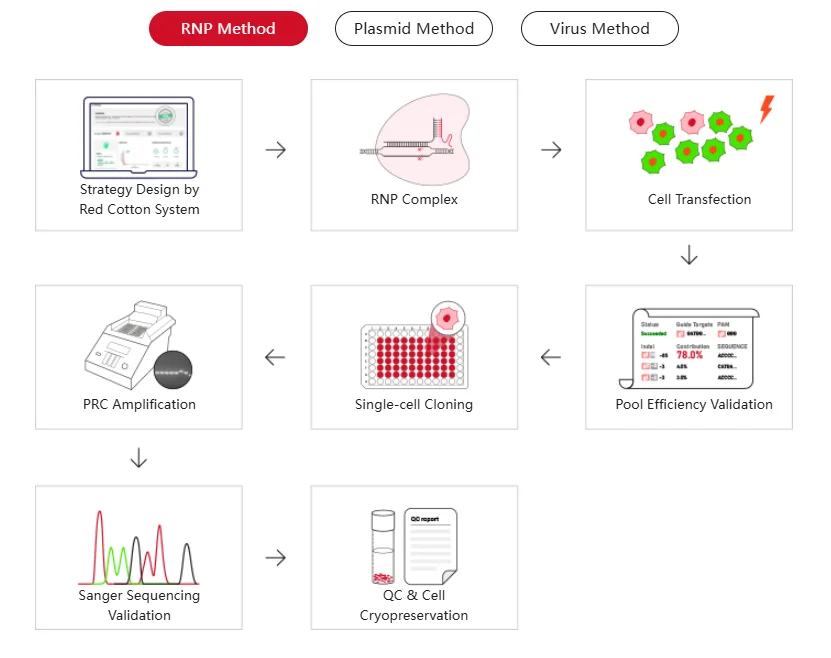
Figure 1. Workflow for the Construction of Stable Knockout Cell Lines Using CRISPR/Cas9.
Experimental Procedures
Step 1. Design and Synthesis of sgRNA and Genotyping Primers
- · sgRNA Design: The first step in generating a knockout cell line is to analyze the target gene sequence and select optimal sgRNA target sites. It is generally recommended to design sgRNAs targeting exons encoding critical functional domains, as this increases the likelihood of achieving complete gene disruption. Key factors to consider during sgRNA design include:
- · On-target editing efficiency score
- · Potential off-target effects, which can be evaluated using design tools such as CRISPOR, Benchling, or Ubigene’s Red Cotton CRISPR Gene Editing System
- · Exon location, reading frame impact, and PAM site accessibility, ensuring efficient and specific targeting
- · Primer Design:PCR primers should be designed to flank the sgRNA target site, typically generating an amplicon of 250–800 bp, suitable for downstream mutation analysis. These primers will be used for detection methods such as T7E1 mismatch cleavage assays or Sanger sequencing to verify genome editing events.
Step 2. Plasmid Construction
The selected sgRNA sequence is cloned into a vector containing Cas9 and a selectable marker (e.g., puromycin or EGFP). Ubigene’s Red Cotton™ gRNA Plasmid Bank includes over 10,000 ready-to-use plasmids, featuring optimized vector backbones that significantly enhance sgRNA expression and Cas9 cleavage efficiency. This platform is particularly suitable for difficult-to-transfect cell types and multi-target genome editing projects, and has been widely applied in research areas including oncology, neuroscience, metabolic diseases, and cell therapy.
Step 3. Plasmid Validation
Sequencing is performed to verify the correct insertion of the sgRNA and confirm sequence integrity, ensuring the absence of mutations, deletions, or incorrect orientation. Plasmid validation is a critical quality control step that prevents false-negative or off-target editing results.
Step 4. Plasmid Preparation and Quality Assessment
High-purity, endotoxin-free plasmids are prepared for transfection. Plasmid concentration and purity are measured (A260/A280 ratio of 1.8–2.0), and integrity is confirmed by agarose gel electrophoresis. Endotoxin contamination or low DNA purity can negatively affect cell viability and transfection efficiency, potentially compromising editing outcomes.
Step 5. Pre-Transfection Optimization
To ensure optimal editing efficiency, the following pre-transfection conditions should be confirmed:
- · Cells are in the logarithmic growth phaseand free from mycoplasma contamination.
- · Cell density is suitable, typically 70–90% confluence.
- · Transfection method is selected based on cell type: chemical transfection, electroporation, or viral delivery.
- · For difficult-to-transfect cells, optimized delivery systems may be required.
Step 6. Cell Transfection/Infection
The CRISPR/Cas9-sgRNA plasmid is introduced into target cells. Appropriate controls should be included:
- · Negative control:empty vector
- · Positive control: a gene with a known, observable phenotype
Transfection efficiency is monitored via fluorescence or antibiotic selection.
Step 7. Cell Pool Screening and Genotyping
48–72 hours post-transfection, the cell pool is collected and genomic DNA is extracted for mutation analysis. Common methods include:
- · PCR + Sanger sequencing:precise identification of insertions/deletions (indels)
- · T7E1 or Surveyor mismatch assays: assessment of editing efficiency and mutation frequency
Ubigene applies a dual PCR + sequencing test to evaluate editing efficiency and selects the most efficiently edited cell pools for downstream single-clone isolation. Using Ubigene’s Genotype Analysis System, sequencing results can be automatically deconvoluted and analyzed, providing rapid and accurate quantification of cell pool knockout efficiency.
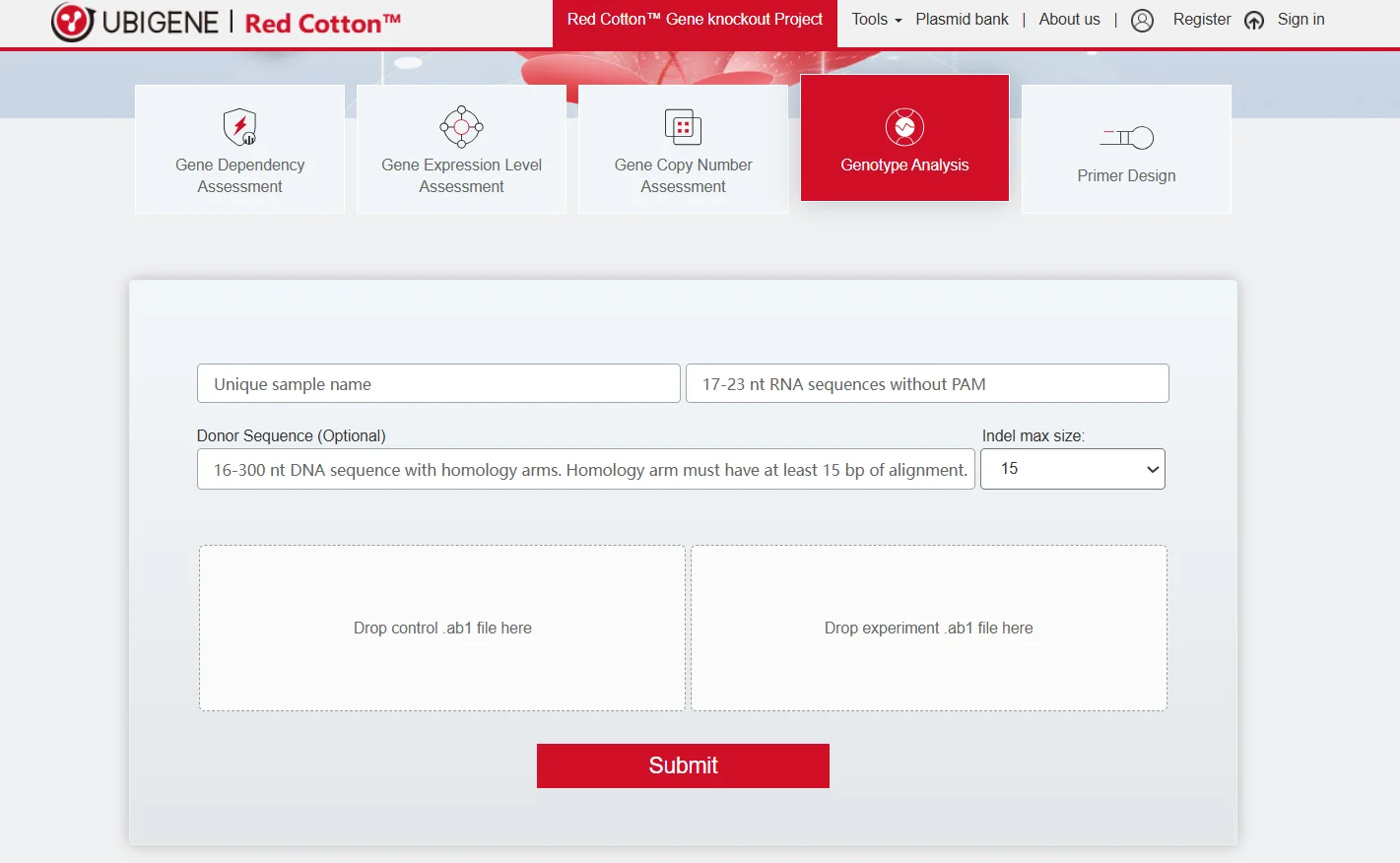
Figure 2. Ubigene Genotype Analysis System.
Step 8. Single-Clone Screening and Expansion
Edited cells are distributed into single wells using either limiting dilution or fluorescence-activated cell sorting (FACS) to establish monoclonal cultures. After expansion, genomic DNA is extracted from each clone for mutation verification to confirm the knockout genotype (homozygous or bi-allelic KO). Verified single clones should undergo functional validation, such as assessing protein expression loss or pathway alterations, to ensure that the gene knockout is effective and phenotypically relevant.
Step 9. Quality Control and Cryopreservation
Once a stable knockout cell line is established, the following quality control steps should be performed:
- · Mycoplasma testing:to confirm absence of contamination
- · STR profiling:to verify cell line identity and authenticity
- · Functional validation:to confirm target gene disruption and assess relevant phenotypes
Cells that pass quality control should be cryopreserved promptly, typically in FBS containing 10% DMSO, to ensure long-term stability and availability for future experiments.
Ubigene Knockout Cell Line Services
Establishing stable knockout cell lines is a technically demanding process that requires extensive expertise and careful consideration of cell line characteristics. Ubigene provides a comprehensive, end-to-end gene knockout solution, assisting researchers in efficiently generating reliable model systems. Key advantages of Ubigene’s services include:
- · Developed Technology Platform: Increases editing efficiency by 10–20-fold, with success rates exceeding 90%in complex cell types.
- · Red Cotton CRISPR Gene Editing System:AI-driven design platform utilizing a database of 1,000+ cell line parameters, enabling automated experimental design and risk assessment.
- · 8000+ Standardized Knockout Cell Linein stock: Ready-to-use cell models that support research across multiple fields.
- · Comprehensive Quality Control System: Includes STR profiling, mycoplasma testing, and functional validation to ensure cell line stability and reliability.
With a track record of over 8,000 successful projects, Ubigene has provided high-quality knockout cell lines to research institutions and pharmaceutical companies worldwide, significantly reducing R&D timelines and facilitating reproducible results.
Conclusion
CRISPR/Cas9-mediated generation of stable knockout cell lines is a critical approach for elucidating gene function and disease mechanisms. Through rational sgRNA design, optimized transfection strategies, and stringent screening and validation, researchers can obtain high-quality, reproducible knockout models. For laboratories seeking to maximize efficiency and success while maintaining experimental control, Ubigene’s professional knockout cell line services provide a comprehensive, reliable, and time-saving solution, supporting accelerated progress in life science research and innovation.



