IF=14.9 | LOTI Gene Editing: Efficient & Precise In Vivo Knock-In


Introduction
The CRISPR–Cas9 system has ushered in a revolutionary era for gene therapy. However, achieving efficient, precise, and low off-target integration of large DNA fragments in somatic cells remains a critical challenge in the field. In a recent study published in Trends in Biotechnology (IF = 14.9), a joint research team from Hainan University and Huazhong University of Science and Technology reported an innovative gene editing strategy termed Long-Offset Paired Nicking–based Target Integration (LOTI) . This strategy employs a Cas9 nickase to introduce long-offset paired nicks in both the genomic target locus and the donor DNA template. Using LOTI, the researchers achieved highly efficient and precise targeted gene knock-in in mouse liver in vivo. Importantly, in a hemophilia B mouse model, LOTI-mediated integration restored up to 55% of normal coagulation factor IX activity, demonstrating strong therapeutic efficacy.
Collectively, these findings establish LOTI as a highly efficient, precise, and safer approach for in vivo targeted integration of large DNA fragments, providing a promising new framework for next-generation gene therapy applications. The development and validation of this innovative gene editing strategy were strongly supported by the Ubigene Monoclone Genotype Validation Kit (extraction free) , which enabled rapid, accurate genotyping and clone validation, facilitating the efficient optimization of advanced gene editing workflows.

Research Background
CRISPR–Cas9–based gene editing technologies have achieved substantial success in gene knockout and small-fragment sequence correction. However, targeted integration of large DNA fragments in somatic cells remains a major bottleneck due to low efficiency, elevated off-target activity, and frequent random insertions.
Conventional strategies predominantly rely on double-strand break (DSB)–induced homology-directed repair (HDR). These approaches exhibit limited efficiency in non-dividing or slowly proliferating cells and are prone to introducing unintended mutations, including insertions, deletions, and chromosomal rearrangements.
Therefore, the development of a DSB-free integration strategy that simultaneously achieves high efficiency and high precision is of critical importance for advancing gene therapy for inherited diseases.
Research Methods
-
Design of the LOTI Strategy:
The LOTI strategy utilizes a Cas9 nickase to introduce a pair of long-offset single-strand nicks (≥200 bp apart) at the genomic target locus. Correspondingly, paired nicks are designed within both the left and right homology arms as well as the internal region of the donor DNA, generating a coordinated architecture of dual genomic nicks combined with triple donor nicks. -
Cell Line and Animal Models:
Knock-in efficiency was evaluated across multiple cell types, including B16, HEK293, and HeLa cell lines, as well as primary astrocytes. Cell cycle inhibitors were applied to assess editing performance in non-proliferating cells. For in vivo studies, a hemophilia B mouse model was established, and gene editing components were delivered to the liver via hydrodynamic tail vein injection. -
Efficiency and Safety Assessment:
Editing outcomes were comprehensively evaluated using flow cytometry, immunofluorescence microscopy, and deep sequencing, enabling quantitative analysis of knock-in efficiency, off-target effects, insertion–deletion (indel) formation, and random genomic integration. -
Mechanistic Investigation:
Key DNA repair pathways involved in LOTI-mediated integration were dissected through siRNA-mediated gene knockdown and small-molecule inhibitors, allowing identification of critical factors required for LOTI-dependent repair processes.
Research Workflow
-
Strategy Construction and Optimization:
The necessity of long-offset paired nicking on both the genomic target site and the donor template was systematically designed and validated. Key parameters, including homology arm length and nick positioning, were further optimized to maximize integration efficiency and precision. -
In Vitro Efficiency Validation:
LOTI was benchmarked against conventional integration strategies, including homology-directed repair (HDR), homology-mediated end joining (HMEJ), and microhomology-mediated end joining (MMEJ), across multiple cell lines to compare knock-in efficiency and targeting accuracy. -
In Vivo Editing Validation:
The efficiency of LOTI-mediated targeted integration was evaluated in mouse liver in vivo, accompanied by comprehensive assessments of off-target activity and overall safety profiles. -
Mechanistic Elucidation:
LOTI-mediated integration was shown to depend on Rad51, Rad52, and DNA ligases I/III, rather than classical non-homologous end joining (NHEJ) or canonical HDR pathways, revealing a distinct DNA repair mechanism underlying LOTI. -
Therapeutic Application in Disease Models:
Targeted integration of the factor IX (FIX) gene was achieved in a hemophilia B mouse model, resulting in restoration of coagulation function and validating the therapeutic potential of LOTI for genetic disease treatment.
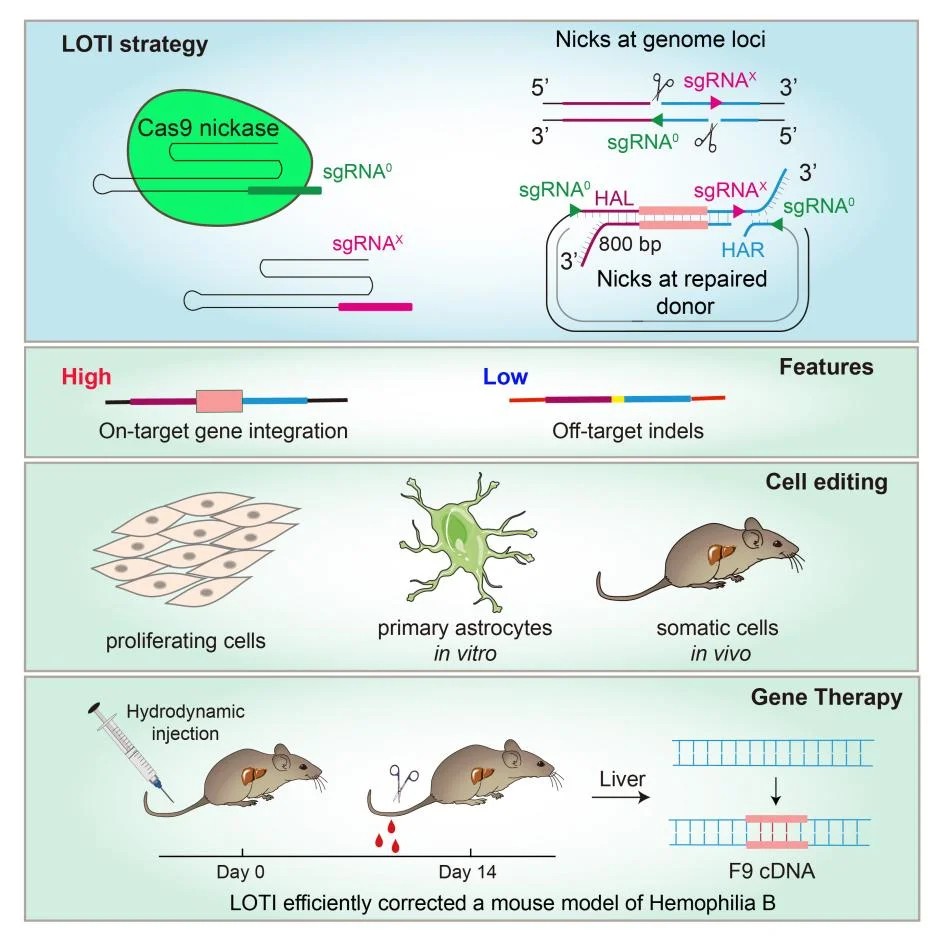
LOTI Strategy and Its Applications
Key Results
-
LOTI Significantly Enhances Knock-In Efficiency in Somatic Cells:
Across multiple cell lines and in mouse liver in vivo, LOTI achieved substantially higher targeted knock-in efficiency compared with conventional single-nick or double-strand break–based strategies. Notably, the long-offset paired nicking design (≥200 bp) was identified as a critical determinant of this enhanced performance, highlighting the importance of spatially separated nicks in promoting efficient and precise integration.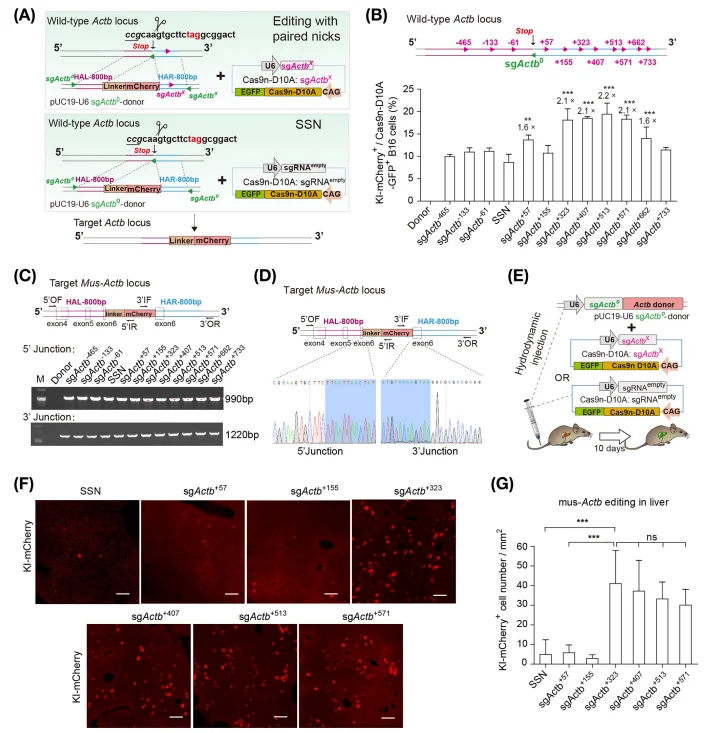
Figure 1. Evaluation of LOTI-Mediated Gene Editing Efficiency In Vitro and In Vivo
-
High Efficiency with Substantially Reduced Off-Target Effects:
In terms of editing accuracy and genomic safety, the LOTI strategy markedly outperformed Cas9 double-strand break–based approaches. LOTI significantly reduced indel frequencies at the on-target locus and minimized random integration of donor DNA. Importantly, at identified off-target sites, LOTI decreased indel formation by approximately 10–100-fold compared with DSB-based Cas9 strategies, demonstrating a substantially improved specificity and safety profile for both in vitro and in vivo gene editing applications.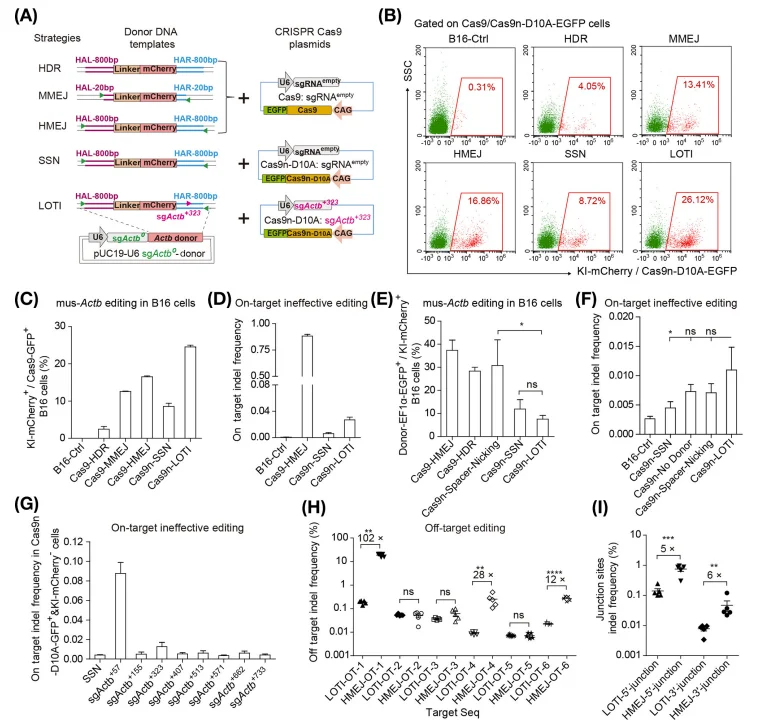
Figure 2. Off-Target Analysis of the LOTI Strategy
-
Broad Applicability to Non-Proliferating Cells and Other Cas9 Variants:
In addition to its robust performance in murine B16 cells, the LOTI strategy demonstrated efficient targeted knock-in in multiple human cell lines, including HEK293 and HeLa cells, as well as in cell cycle–arrested cells and primary mouse astrocytes. Moreover, LOTI remained effective when implemented with the SaCas9-D10A nickase, suggesting that the LOTI framework is compatible with alternative CRISPR–Cas nickase variants and may be broadly applicable across different CRISPR nickase systems.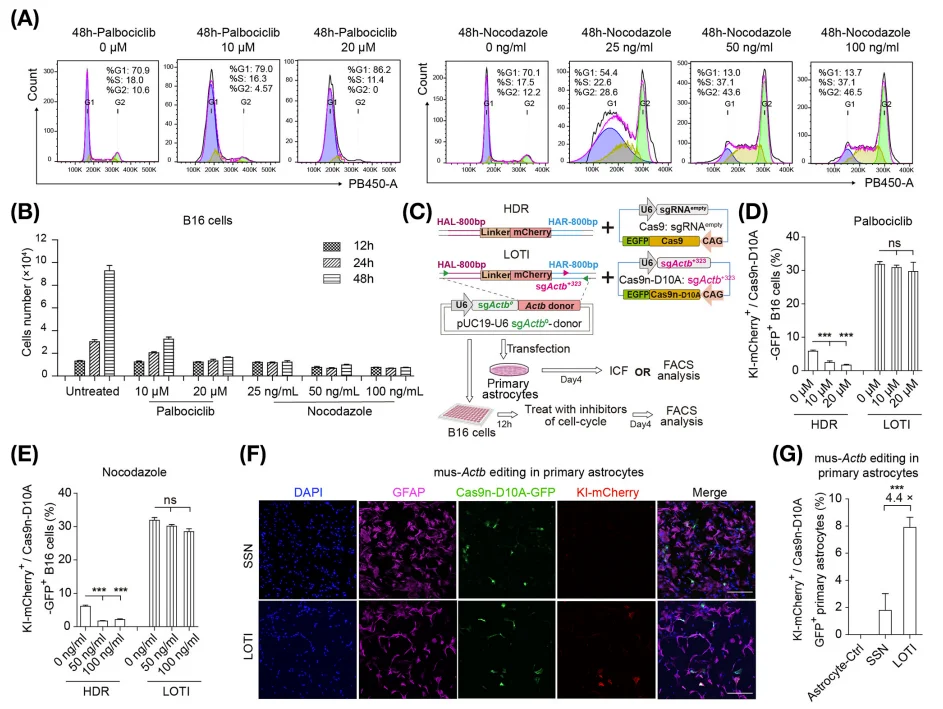
Figure 3. Performance of the LOTI Strategy in Cell Cycle–Arrested Cells and Primary Astrocytes
-
Effective Treatment of a Hemophilia B Mouse Model:
Hemophilia B is an inherited bleeding disorder caused by mutations or deletions in the coagulation factor IX (FIX) gene, resulting in impaired blood coagulation. To evaluate the therapeutic potential of LOTI, a LOTI-mediated targeted integration strategy was designed for the FIX locus. The engineered editing components were delivered in vivo to a hemophilia B mouse model via hydrodynamic tail vein injection. A single administration of LOTI components achieved stable targeted integration of the FIX gene in the liver of F9-knockout mice, leading to restoration of up to 55% of normal coagulation activity. Consistently, both FIX mRNA and protein expression levels were robustly restored, demonstrating effective functional correction and highlighting the strong therapeutic potential of LOTI for the treatment of inherited coagulation disorders.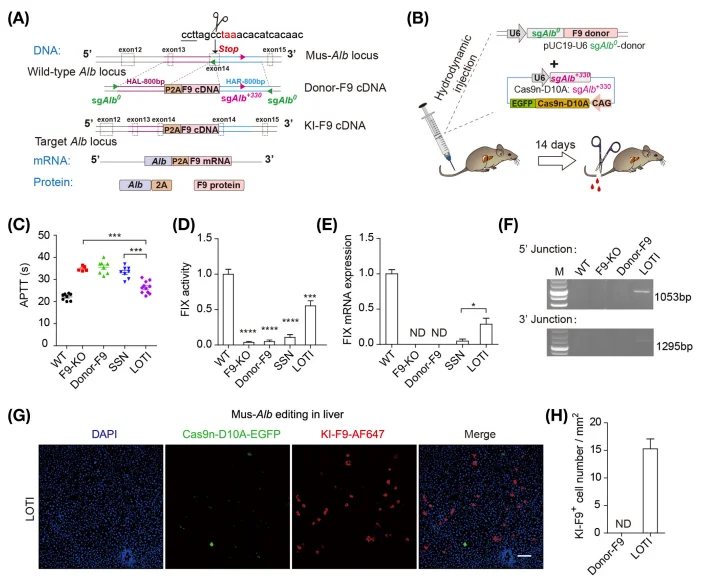
Figure 4. Application of the LOTI Strategy in a Hemophilia B Mouse Model
Significance and Innovations
-
Strategic Innovation:
This study is the first to introduce a coordinated design combining long-offset paired nicking with multiple donor nicks, enabling efficient and precise large-fragment gene integration in somatic cells. This approach overcomes the fundamental limitations of conventional genome editing strategies that rely on double-strand breaks. -
Novel Mechanistic Insights:
The study reveals that LOTI activates a Rad51/Rad52-dependent non-canonical DNA repair pathway, operating independently of the cell cycle. These findings provide new mechanistic insights into nick-guided targeted gene integration. -
Therapeutic Breakthrough:
In a hemophilia B mouse model, LOTI enabled robust gene correction following a single in vivo administration, demonstrating a safe and effective strategy for the treatment of genetic diseases caused by large gene deletions. -
Platform Potential:
With its multi-nuclease compatibility, high precision, and low off-target activity, LOTI represents a scalable and broadly applicable genome editing platform with potential applications across genetic disorders, metabolic diseases, and cancer gene therapy.
Summary
In this study, the LOTI strategy was developed based on a synergistic design of long-offset paired genomic nicks and multiple donor nicks, enabling efficient, precise, and low off-target integration of large DNA fragments in somatic cells. The therapeutic efficacy of LOTI was further validated in a hemophilia B mouse model, establishing a new framework for the development of advanced genome editing technologies.
Support Provided by Ubigene
The development of this novel gene editing strategy was strongly supported by the Ubigene Monoclone Genotype Validation Kit (extraction free) . This kit enables early-stage genotyping of monoclonal cells, allowing rapid identification of correctly edited clones and shortening experimental timelines by 2–4 weeks.
Leveraging over 15 years of proprietary expertise, Ubigene has developed the EZ-editor™ Gene Editing Product Series , comprising four integrated product lines designed to simplify workflows, enhance efficiency, and deliver reliable editing outcomes. With a strong focus on detail-oriented innovation, Ubigene empowers researchers to make gene editing faster, easier, and more precise.
Contact us to learn more >>>Reference
Lu Y, Wang J, Xu Y, Xu M, Li B, Fan Z, Liu J, Li X, Cai Z, Zheng Y, Wang W, Yang J, Zhang Z, Liu Z. Long-offset paired nicking-based efficient and precise strategy for in vivo targeted insertion. Trends Biotechnol. 2025 Jul;43(7):1743-1764. doi: 10.1016/j.tibtech.2025.02.020. Epub 2025 Apr 7. PMID: 40199626.


