CRISPR Screen Identifies Autophagy-Related Gene as “Culprit” in Liver Cancer Drug Resistance


CRISPR Screen Identifies Autophagy-Related Gene as “Culprit” in Liver Cancer Drug Resistance
Introduction
Autophagy is a cellular self-cleaning process that degrades damaged organelles and proteins to maintain cell health. In recent years, the role of autophagy in cancer has received increasing attention, especially in tumor drug resistance. Recently, a research published in Autophagy titled “Genome-Scale CRISPR screen identifies LAPTM5 driving lenvatinib resistance in hepatocellular carcinoma” used CRISPR library to perform a genome-wide screen in liver cancer cells, identifying a key autophagy-related gene: LAPTM5. Subsequently, HCC cell lines, patient-derived primary cell lines, human HCC xenografts, and immunocompetent mouse HCC models were used to verify that LAPTM5 mediates resistance to lenvatinib in liver cancer by regulating the autophagy process.
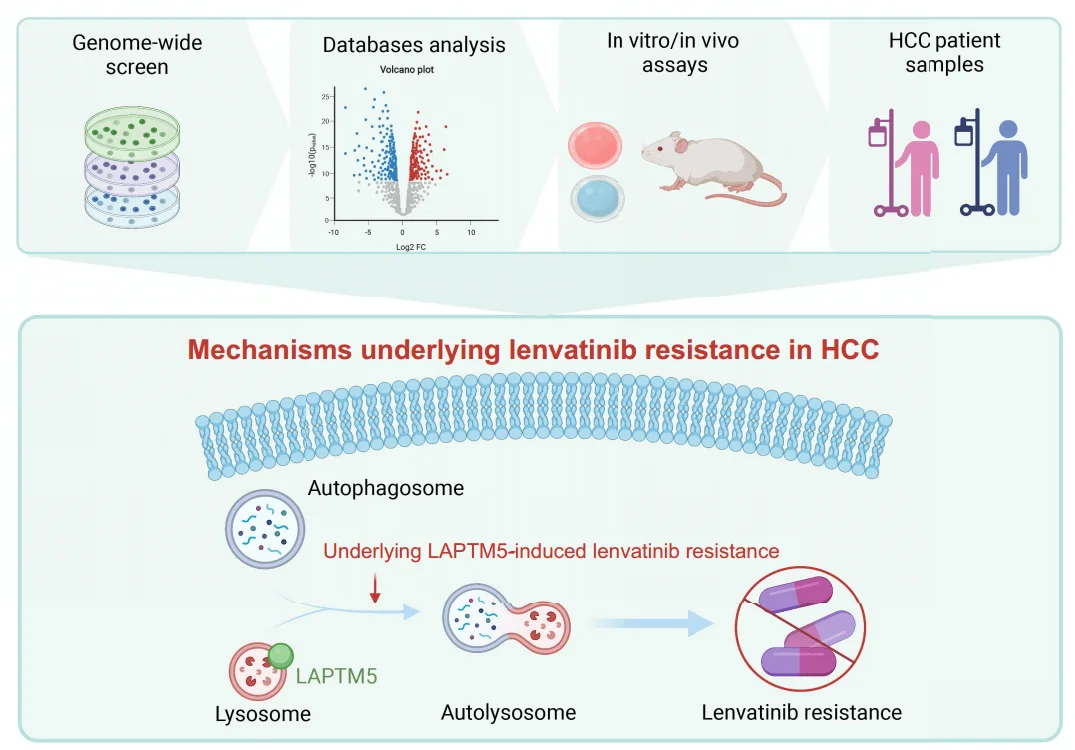
Graphical Abstract
Today, we will interpret this study from the perspective of autophagy research to see how the CRISPR library helps scientists discover key factors in drug resistance mechanisms!
The researchers used CRISPR-Cas9 library screening technology to perform genome-wide gene knockout screening of liver cancer cells to find out which gene deletions would affect the sensitivity of cells to lenvatinib. Integrated genome-wide screening and database analysis identified LAPTM5 and TNXB as potential factors in lenvatinib resistance in HCC.
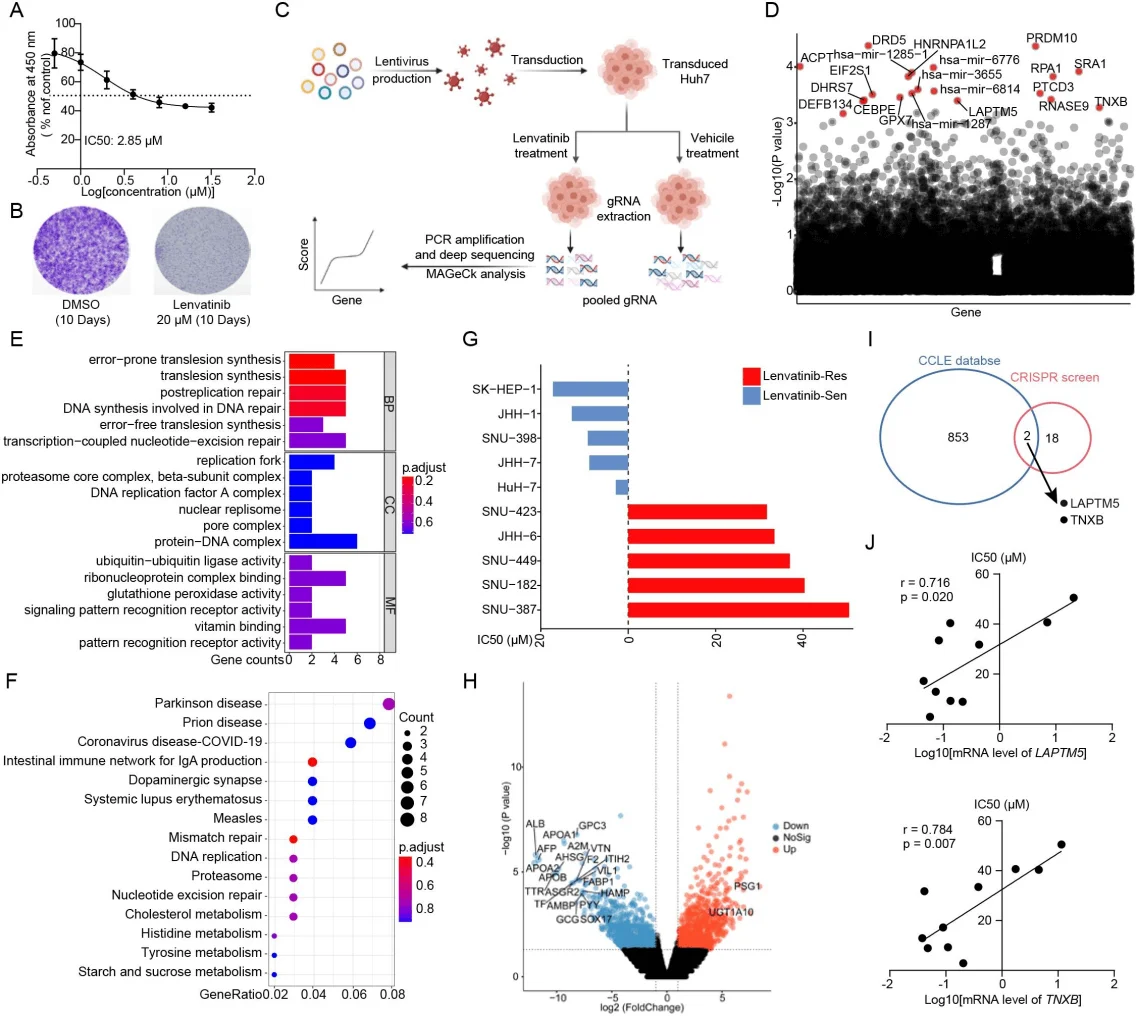
Integrating genome-wide CRISPR-Cas9 screening and database analysis reveals key factors for lenvatinib resistance
To verify the effects of TNXB and LAPTM5 on drug resistance, their expression levels in HCC and normal liver tissues were compared by analyzing TCGA and GTEx databases. si-knockdown cells, sh-interference stable cell lines, and overexpression stable cell lines were constructed for in vitro drug resistance-related studies. The results showed that deletion of the LAPTM5 gene significantly enhanced the sensitivity of liver cancer cells to lenvatinib, while its overexpression made the cells resistant.
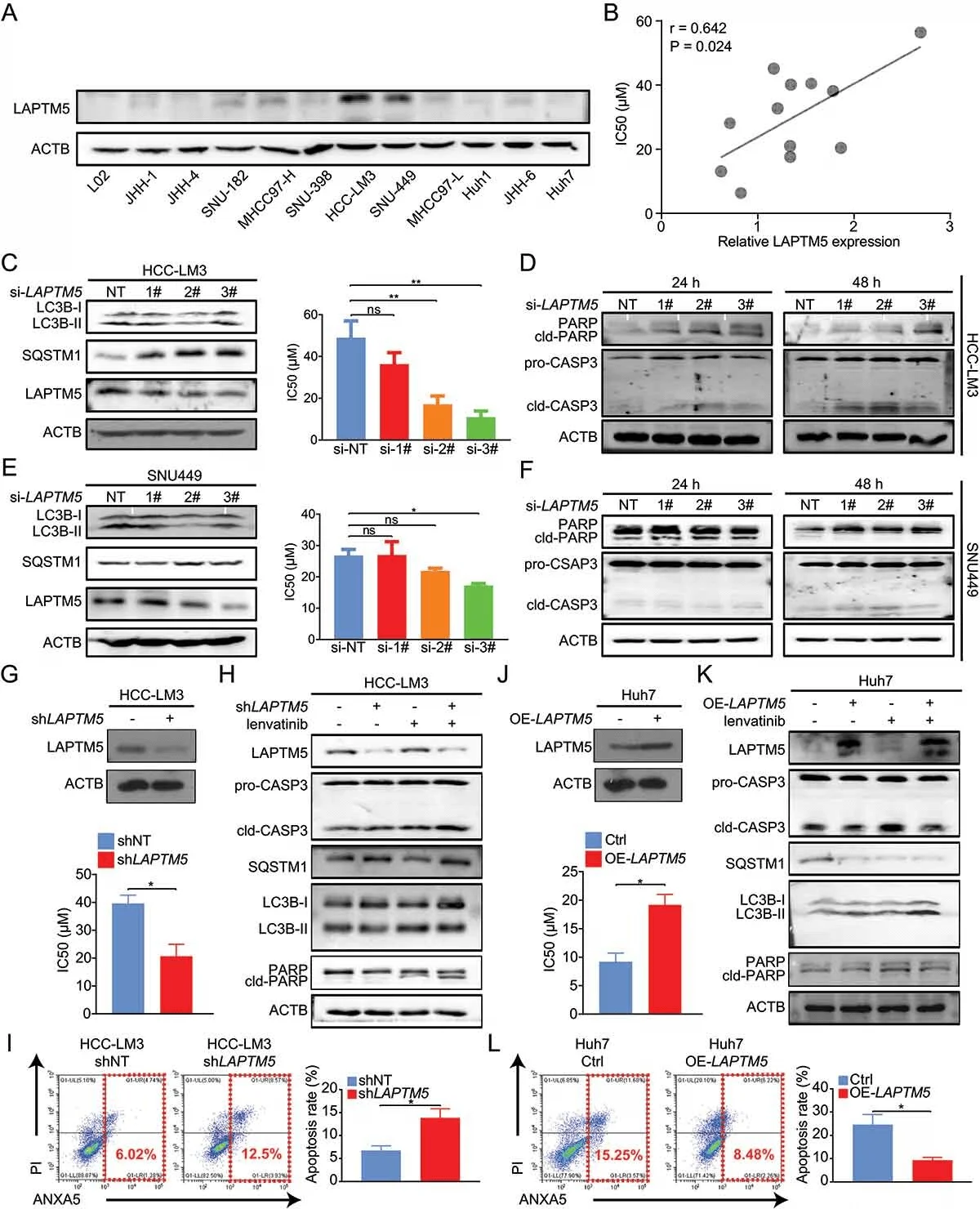
LAPTM5 contributes to lenvatinib resistance in HCCin vitro
Then, researchers explored the relationship between LAPTM5 and the sensitivity of HCC to lenvatinib through in vivo mouse experiments. The results were consistent with in vitro cell experiments. In vitro and in vivo experiments showed that LAPTM5 plays a key role in driving lenvatinib resistance in HCC.
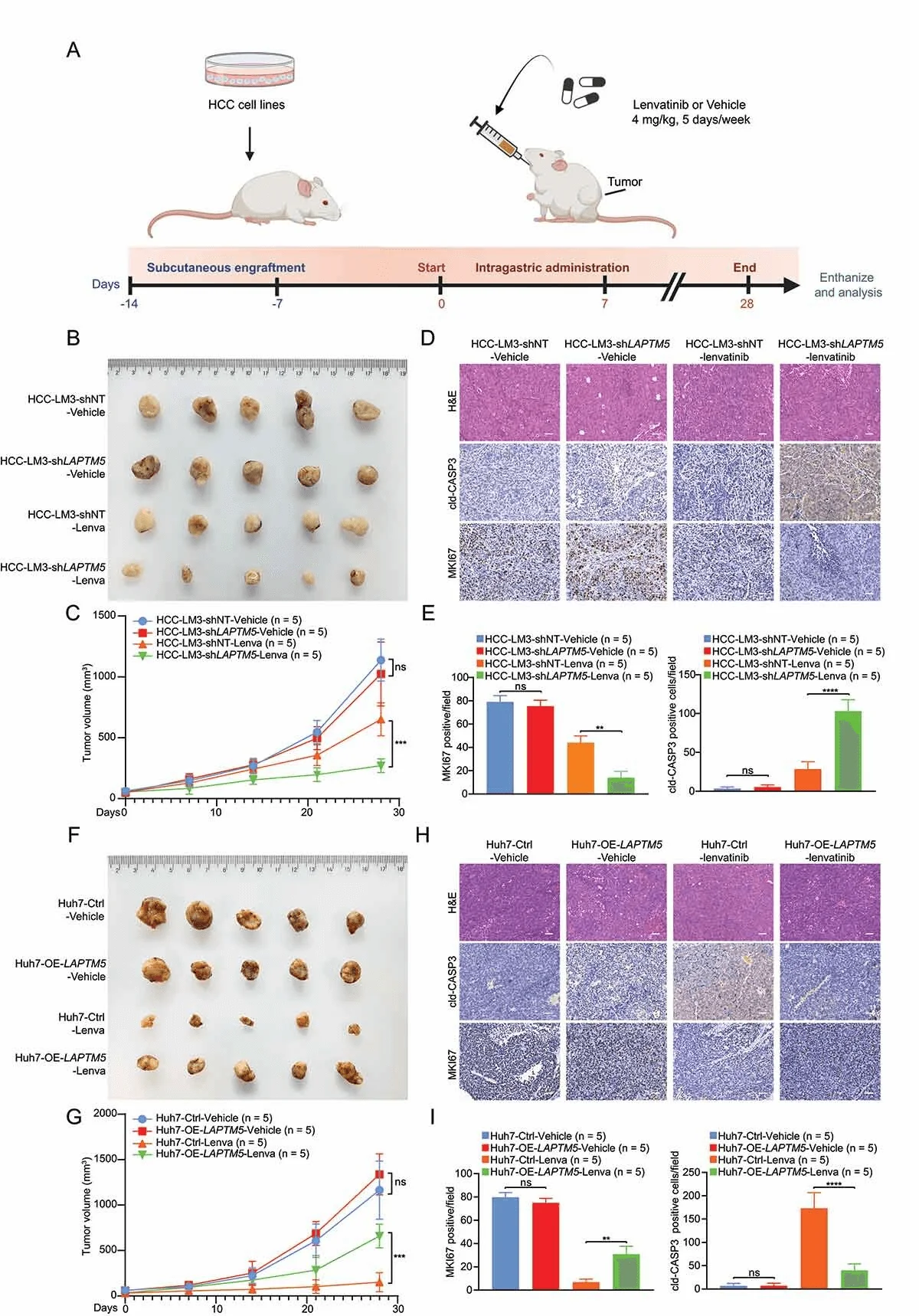
LAPTM5 causes lenvatinib resistance in HCC in vivo
To understand how LAPTM5 causes drug resistance, researchers further studied its mechanism of action and found that LAPTM is closely related to the autophagy process. LAPTM5 can inhibit the function of lysosomes. Lysosomes are key organelles in the autophagy process, responsible for degrading damaged organelles and exogenous substances (such as drugs). When LAPTM5 is excessive, lysosome function is inhibited, the autophagy process is blocked, and lenvatinib cannot be effectively degraded, allowing cancer cells to escape. In addition to directly inhibiting lysosomal function, LAPTM5 also affects the autophagy process by regulating the mTOR signaling pathway. mTOR is a cellular nutrient sensor that can inhibit autophagy. Studies have found that LAPTM5 overexpression activates the mTOR signaling pathway, thereby inhibiting autophagy and further promoting cancer cell survival and drug resistance.
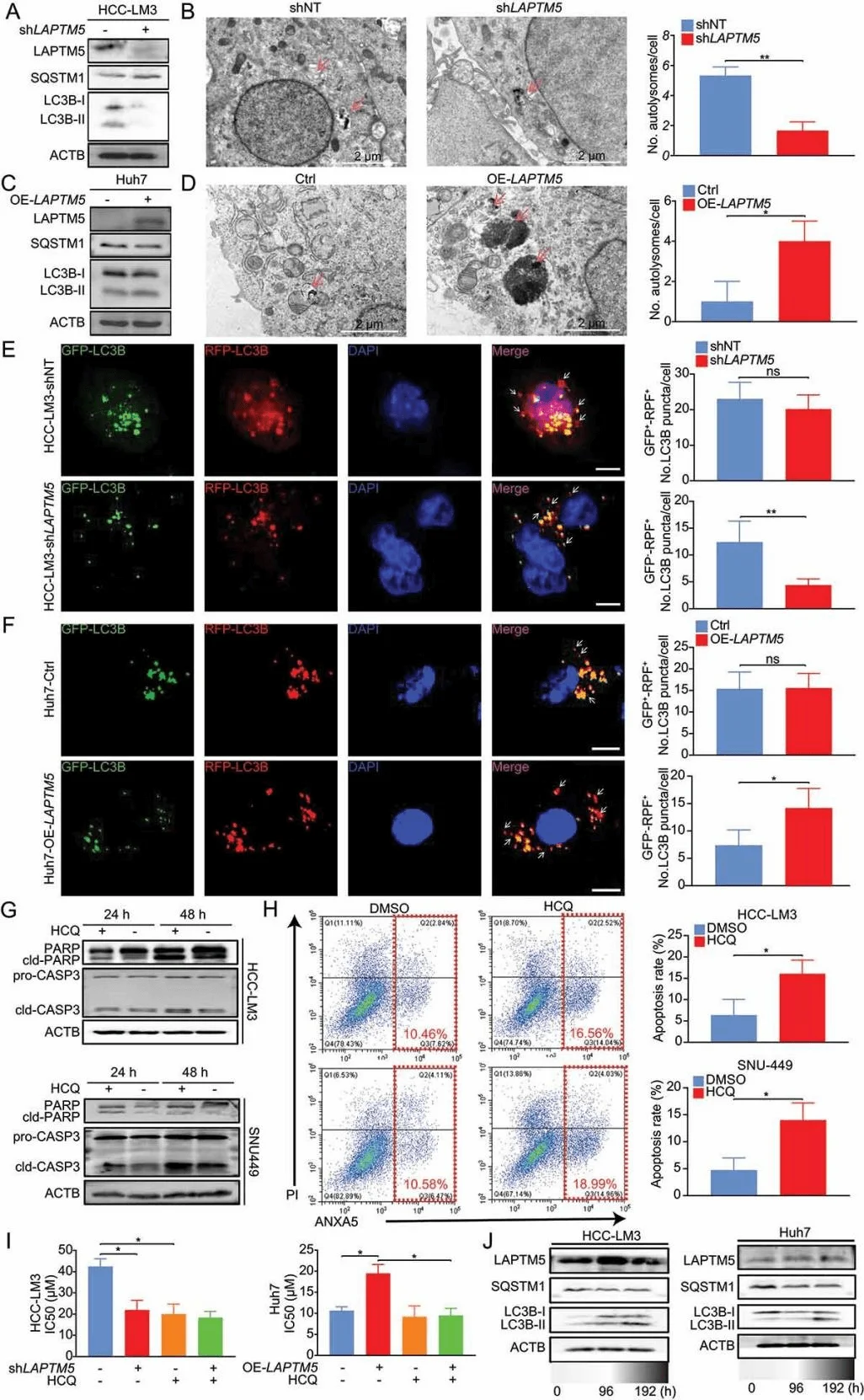
LAPTM5 enhances autophagic flux to reduce HCC sensitivity to lenvatinib
Considering that HCC is a heterogeneous tumor with different genetic backgrounds, researchers also studied the effects of LAPTM5 on lenvatinib resistance using PDC and PDO models. The results showed that LAPTM5 is a key factor in lenvatinib resistance in HCC, regardless of genetic background.
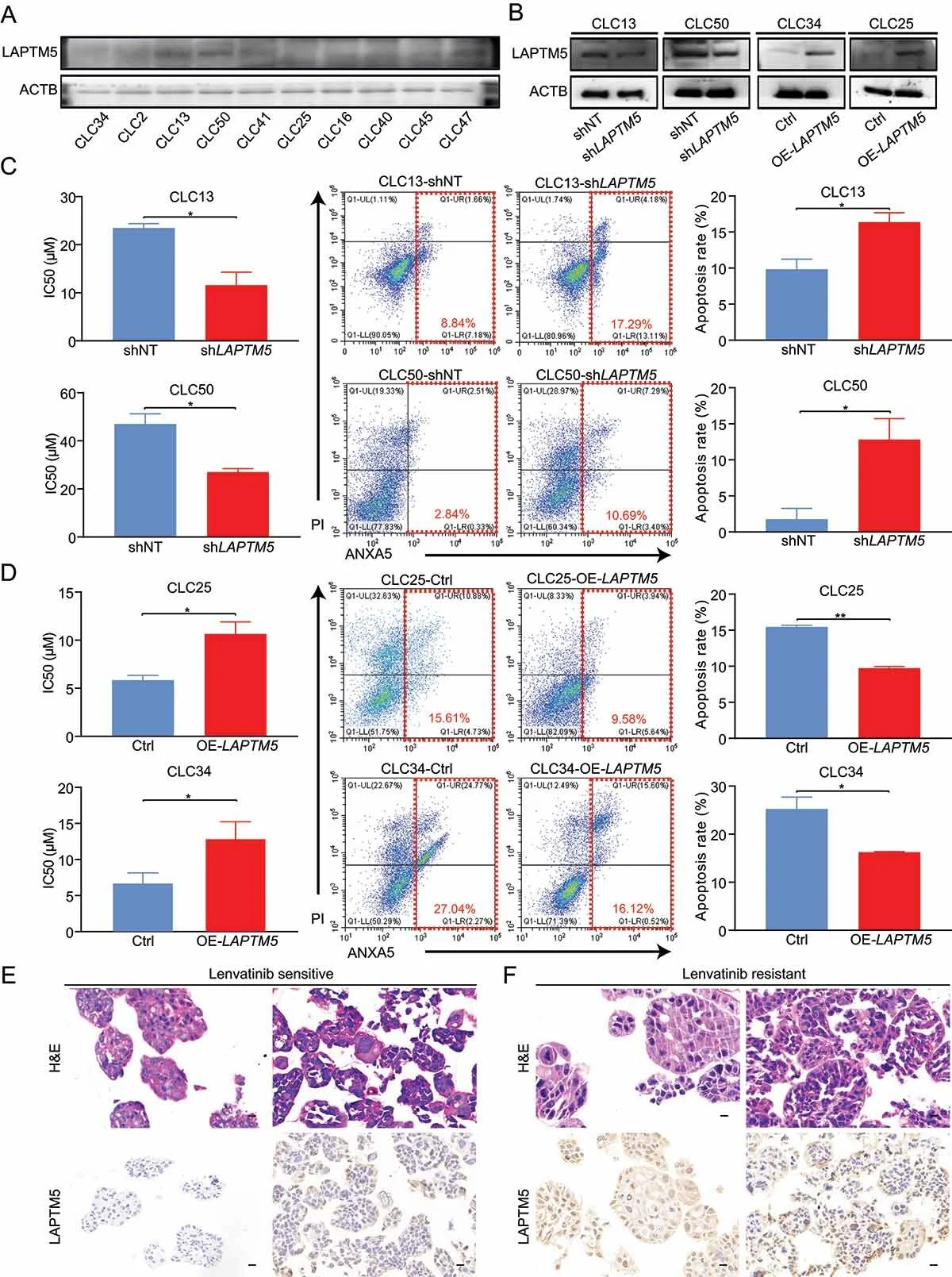
PDC and PDO models analyze the correlation between LAPTM5 and HCC sensitivity to lenvatinib
To further verify the clinical significance of LAPTM5, researchers then assessed the association between LAPTM5 and lenvatinib resistance by analyzing clinical samples. The researchers found that the level of LAPTM5 expressed in recurrent patients was significantly higher than in non-recurrent patients (1.87 times, P < 0.001). This observation indicates that LAPTM5 upregulation leads to lenvatinib resistance, as early recurrence after lenvatinib treatment may also be considered a sign of resistance. LAPTM5 may serve as a reliable biomarker for predicting patient response to lenvatinib, thereby providing precise guidance for clinical intervention.
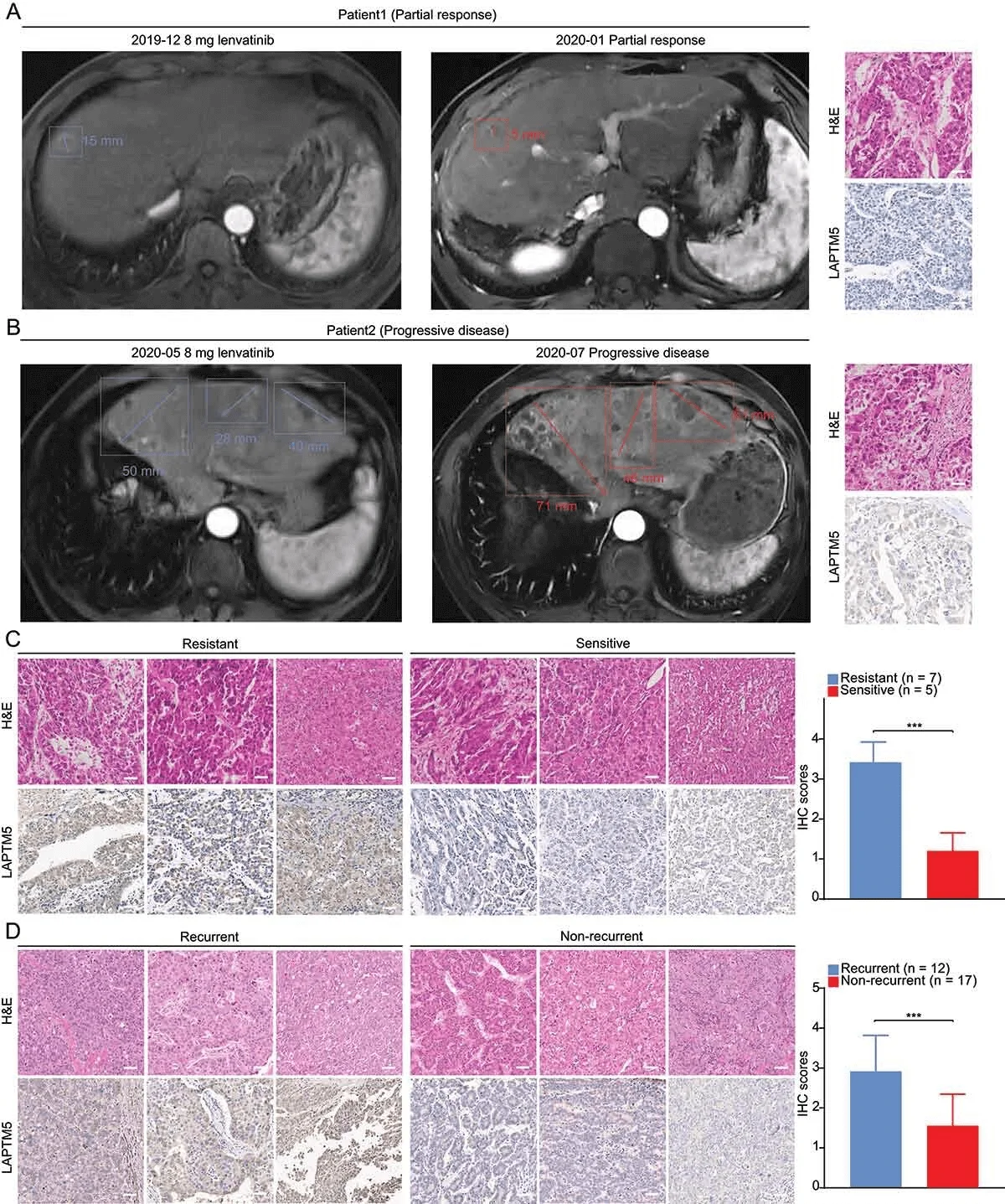
Evaluation of the association between LAPTM5 and response to lenvatinib in clinical samples
Summary
This study used CRISPR screening technology to reveal the key role of LAPTM5 in lenvatinib resistance in liver cancer. LAPTM5 helps cancer cells escape drug attacks by inhibiting the autophagy process. This discovery not only provides a new perspective for understanding drug resistance mechanisms but also provides potential new strategies for future liver cancer treatment.
The Importance of CRISPR Screening in Autophagy Research:
1.High-Throughput Screening: CRISPR libraries can quickly and efficiently screen for genes related to autophagy, providing a powerful tool for studying autophagy mechanisms.
2.Precise Regulation: Through CRISPR technology, scientists can precisely knock out or overexpress specific genes to deeply study their role in autophagy.
3.Value of transforming into medical applications: Genes discovered by CRISPR screening (such as LAPTM5) may become new therapeutic targets, bringing hope for cancer treatment.
Enlightenment
1.Hope for Solving Drug Resistance: In the future, drugs targeting LAPTM5 may become a new weapon to overcome lenvatinib resistance.
2.New Direction for Precision Medicine: By detecting the expression level of LAPTM5, doctors may be able to predict the patient's treatment response to lenvatinib and achieve personalized treatment.
Prospect
This study brings new hope for liver cancer treatment, but there is still a long way to go before clinical application. In the future, scientists need to further verify the safety and effectiveness of LAPTM5-targeted therapy to bring more benefits to liver cancer patients.
Ubigene’s CRISPR Library Products - Screening-ready Cell Pool
Standard upgrade, ultra-high coverage
400+ in-stock, delivery fast as 1 week
Ready to use for screening, saving 10 weeks to screen the target! Feel free to contact us!
Reference:


