CRISPR Library In Vitro Screening
CRISPR Library In Vitro Screening refers to the process of applying specific selective pressures to a pre-constructed cell pool, inducing phenotypic changes, and subsequently enriching cells through targeted methods. During screening, cells that acquire increased resistance to the selective pressure through gene editing gradually gain a growth advantage, whereas cells that become more sensitive undergo apoptosis or growth arrest. This “survival of the fittest” process allows for the identification of functional targets by evaluating the abundance of sgRNAs in the post-selection cell population.
Why choose Ubigene?
Ubigene has established a comprehensive phenotypic screening and data analysis platform tailored to various research scenarios. We provide refined functional screening services, covering the entire workflow from experimental design to result interpretation, enabling efficient advancement of scientific research.
Diverse Phenotypic Analysis Platform
Ubigene's diversified phenotypic analysis platform supports a wide range of functional screening systems. It enables comprehensive coverage of various phenotypes, including flow cytometry-based sorting, cell migration, and adherent cell assays, across multiple experimental setups.
Mature and Stable Cell Biology Platform
Our cell biology platform meets large-scale cell culture requirements, with experienced personnel skilled in drug/virus treatment, passaging, and co-culture techniques. This ensures functional screening can be effectively performed across diverse cellular systems.
Expert Technical Team and Project Management
Ubigene has an expert team with extensive knowledge and hands-on experience in functional screening systems. They provide step-by-step guidance for experimental design and operation, ensuring smooth execution of functional screens.
Comprehensive, End-to-End Service
With a robust pre-experiment optimization and quality management system, Ubigene delivers full-process functional screening and one-stop CRISPR screening services. This integrated approach seamlessly connects all workflow stages, ensuring experimental continuity and maximizing success rates.
Construction of CRISPR Library In Vitro Screening System
CRISPR library in vitro screening system is designed based on customized research needs and target phenotypes. A complete functional screening system consists of two key components: “selective pressure” and “phenotypic enrichment.”
Selective Pressure
"Selective Pressure" refers to imposing specific screening pressures on a pre-constructed cell pool according to the experimental objectives. This induces cells with different gene edits to exhibit the desired phenotypes, allowing them to be effectively distinguished during subsequent enrichment stages. Common methods for applying selective pressure include passive approaches, such as serial passaging, or active interventions, such as drug treatment, viral infection, co-culture with functional cells, or stimulation with cytokines.
Serial Passaging
Drug Treatment
Functional Cell Co-Culture
Viral Infection
Cytokine Stimulation
Other Treatments
Phenotypic Enrichment
"Phenotypic Enrichment" refers to selecting appropriate enrichment strategies to collect cells exhibiting the desired phenotypes after the application of selective pressure. Common target phenotypes used to distinguish cell populations include changes in cell proliferation or apoptosis, variations in the expression levels of specific markers, and behavioral phenotypes such as cell migration or adhesion.
Based on Cell Proliferation/Apoptosis
Based on Behavioral Phenotypes
Based on Cell-Specific Marker Expression
Establish a Customized CRISPR Library In Vitro Screening System
Workflow
By leveraging the orthogonal combination of multiple selective pressures and enrichment strategies, Ubigene has developed a flexible and versatile functional screening platform. Clients can customize both the “applied pressure” and “enrichment strategy” according to their specific research objectives, enabling more precise functional discovery.
CRISPR Library Cell Pool Construction
Application of Selective Pressure & Phenotypic Enrichment
Functional screening system
Genomic DNA Extraction
NGS Sequencing
Report & Data Analysis
CRISPR Library In Vitro Screening
CRISPR Library In Vivo Screening

Selective Pressure
Phenotypic Enrichment
Serial Passaging
+Phenotypic Enrichment Based on Cell Proliferation/Apoptosis
Serial Passaging
+Phenotypic Enrichment Based on Cell-Specific Marker Expression
Serial Passaging
+Phenotypic Enrichment Based on Behavioral Phenotypes
Drug Treatment
+Specific Enrichment Strategy
Viral Infection
+Specific Enrichment Strategy
Functional Cell Co-Culture
+Specific Enrichment Strategy
Cytokine Stimulation
+Specific Enrichment Strategy

Typical applications:
- Screening for genes essential for cell growth/survival
- Screening for synthetic lethality targets

Selective
Pressure
Phenotypic
Enrichment
Serial Passaging
+Phenotypic Enrichment Based on Cell Proliferation/Apoptosis

Typical applications:
Screening for genes essential for cell growth/survival
Screening for synthetic lethality targets
Serial Passaging
+Phenotypic Enrichment Based on Cell-Specific Marker Expression

Typical applications:
Antigen epitope recognition
Study of signaling pathway regulatory mechanisms
Study of single-gene regulatory mechanisms
Serial Passaging
+Phenotypic Enrichment Based on Behavioral Phenotypes

Typical applications:
Regulators of cell migration
Regulators of cell adhesion
Drug Treatment
+Specific Enrichment Strategy

Typical applications:
Screening for tumor drug resistance genes
Screening for genes involved in drug synergistic effects
Investigation of drug targets and mechanisms of action
Viral Infection
+Specific Enrichment Strategy

Typical applications:
Identification of host factors related to viral infection
Screening for genes involved in vaccine synergistic effects
Investigation of vaccine targets and mechanisms of action
Functional Cell Co-Culture
+Specific Enrichment Strategy

Typical applications:
Identification of genes regulating immune cell persistence/exhaustion resistance
Identification of genes regulating tumor cell sensitivity to immune killing
Identification of genes regulating immune cell activation-induced cell death
Identification of genes influencing cytokine/nutrient competition for survival advantage
Cytokine Stimulation
+Specific Enrichment Strategy

Typical applications:
Exploration of factors promoting or inhibiting specific cellular biological processes
Study of signaling pathway regulatory mechanisms
Investigation of cytokine targets and mechanisms of action
Representative publications
CRISPR Library In Vitro Screening System: Serial passaging (selective pressure) + cell survival/proliferation (enrichment method)
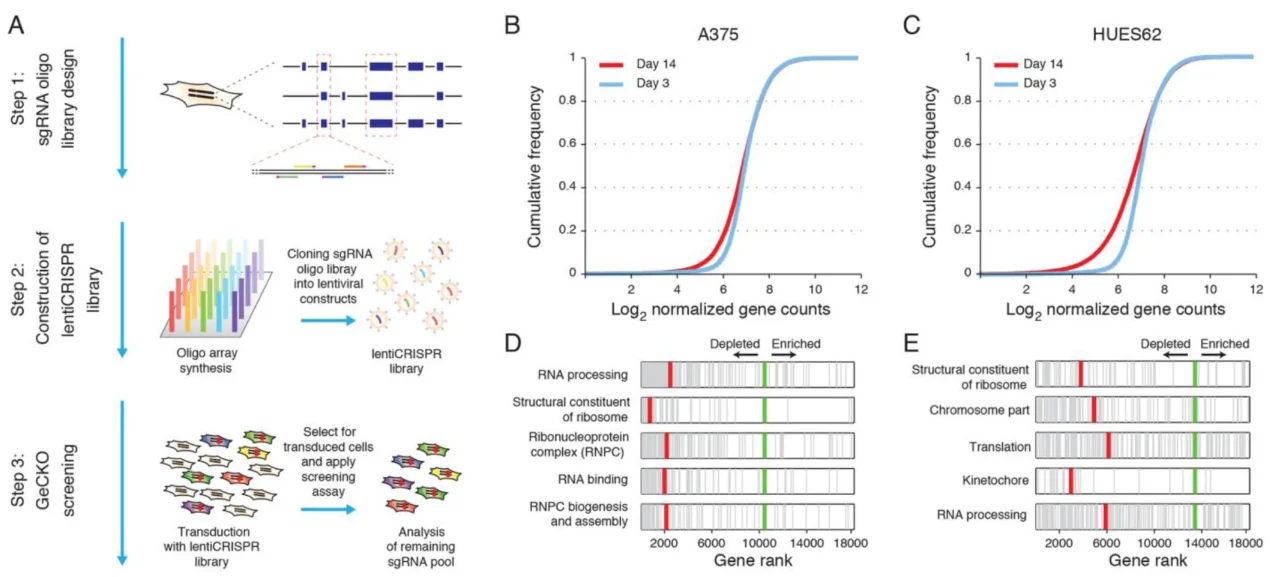
1. CRISPR-Cas9 knockout screening in human cells, Science.
Application: Identification of genes essential for the growth of human melanoma cells and human pluripotent stem cells
Currently, this type of functional screening system is relatively rare. It was widely used in the early development of CRISPR screening systems to identify genes required for growth or proliferation across various cell types. In this study, the authors successfully applied this screening system to identify genes essential for the growth of human melanoma cells and human pluripotent stem cells, providing valuable guidance for advancing CRISPR screening technology and offering a strong theoretical foundation for the field of tumor biology.
View Picture
CRISPR Library In Vitro Screening System: Serial passaging + enrichment based on cell-specific marker expression levels
2. CRISPR screening identifies the deubiquitylase ATXN3 as a PD-L1-positive regulator for tumor immune evasion, J Clin Invest.
Application: Study of single-gene regulatory mechanisms
The authors constructed a CRISPR screening platform covering 96 members of the deubiquitylase family. By analyzing cell populations with low and high PD-L1 expression, they identified ATXN3 as a positive transcriptional regulator of PD-L1. Tumors lacking ATXN3 responded better to low-dose anti-PD-1 treatment, and inhibition of ATXN3 enhanced the efficacy of immune checkpoint blockade therapy.
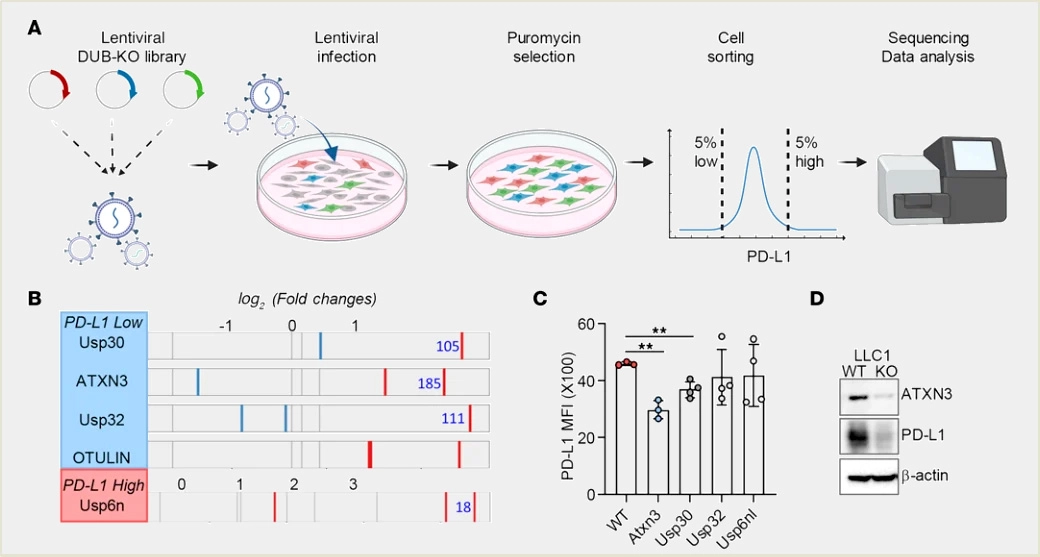
View Picture
CRISPR Library In Vitro Screening System: Drug treatment (selective pressure) + cell survival/proliferation (enrichment method)
3. Gliocidin is a nicotinamide-mimetic prodrug that targets glioblastoma, Nature.
Application: Investigation of the mechanism of action of the anti-glioblastoma compound Gliocidin
In this study, the authors employed a whole-genome mouse CRISPR knockout library (Brie) to perform CRISPR screening in NG2-3112 cells. Cells were treated with Gliocidin at different concentrations (IC50 and IC80) and harvested on Day 0 and Day 14 post-treatment. Results from positive and negative selection screens indicated that positive and negative regulators of mTORC1 respectively decreased and increased tumor cell sensitivity to Gliocidin, highlighting the critical role of the mTORC1 pathway in mediating the anti-glioblastoma efficacy of Gliocidin.
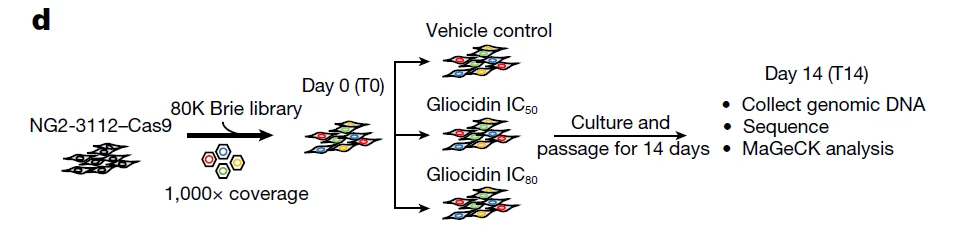
View Picture
CRISPR Library In Vitro Screening System: Drug treatment (selective pressure) + flow cytometry-based enrichment (enrichment method)
4. An E3 ligase network engages GCN1 to promote the degradation of translation factors on stalled ribosomes, Cell.
Application: Investigation of the mechanism by which Ternatin-4 mediates eEF1A degradation
In this study, the authors generated a cell line overexpressing mCherry-eEF1A fusion protein and performed CRISPRi screening of UPS gene sets in this cell line. Based on previous findings that Ternatin-4 promotes eEF1A degradation, they treated the cells with Ternatin-4 and enriched the populations according to mCherry fluorescence intensity. This approach led to the identification of RNF14 and RNF25 as key mediators of eEF1A and ribosomal protein ubiquitination.
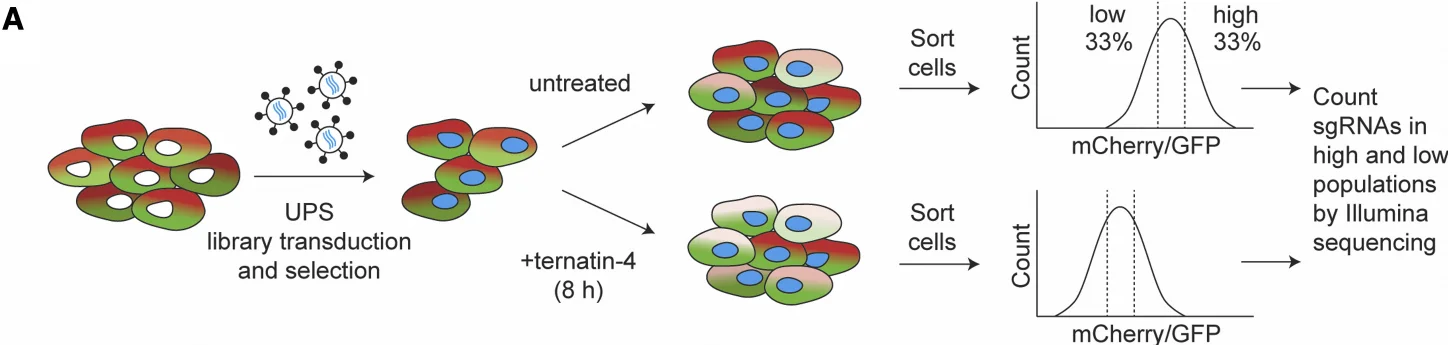
View Picture
CRISPR Library In Vitro Screening System: Drug treatment (selective pressure) + behavioral phenotype-based enrichment (enrichment method)
5. Whole-genome screens reveal regulators of differentiation state and context-dependent migration in human neutrophils, Nature Communications.
Application: Identification of genes involved in adhesion-dependent and -independent cell migration, protein trafficking, and actomyosin cytoskeleton regulation
In this study, the authors used a whole-genome CRISPRi library and designed three experimental models to investigate key regulators associated with different migration behaviors of neutrophils. In two models, cells were seeded in a top reservoir above a track-etched membrane with 3 μm pores. A chemical gradient was established by adding 10% heat-inactivated fetal bovine serum to different regions of the reservoir to assess ordered and disordered chemotactic migration. In the third model, cells were embedded in a synthetic extracellular matrix to study amoeboid 3D migration, simulating movement through tissue interstices. Using these models, the authors identified 344 genes whose knockdown decreased the proportion of migrating cells and 31 genes whose knockdown increased migration. They also revealed the role of mTORC1 signaling in HL60 cell differentiation, affecting neutrophil abundance, survival, and migration behavior.
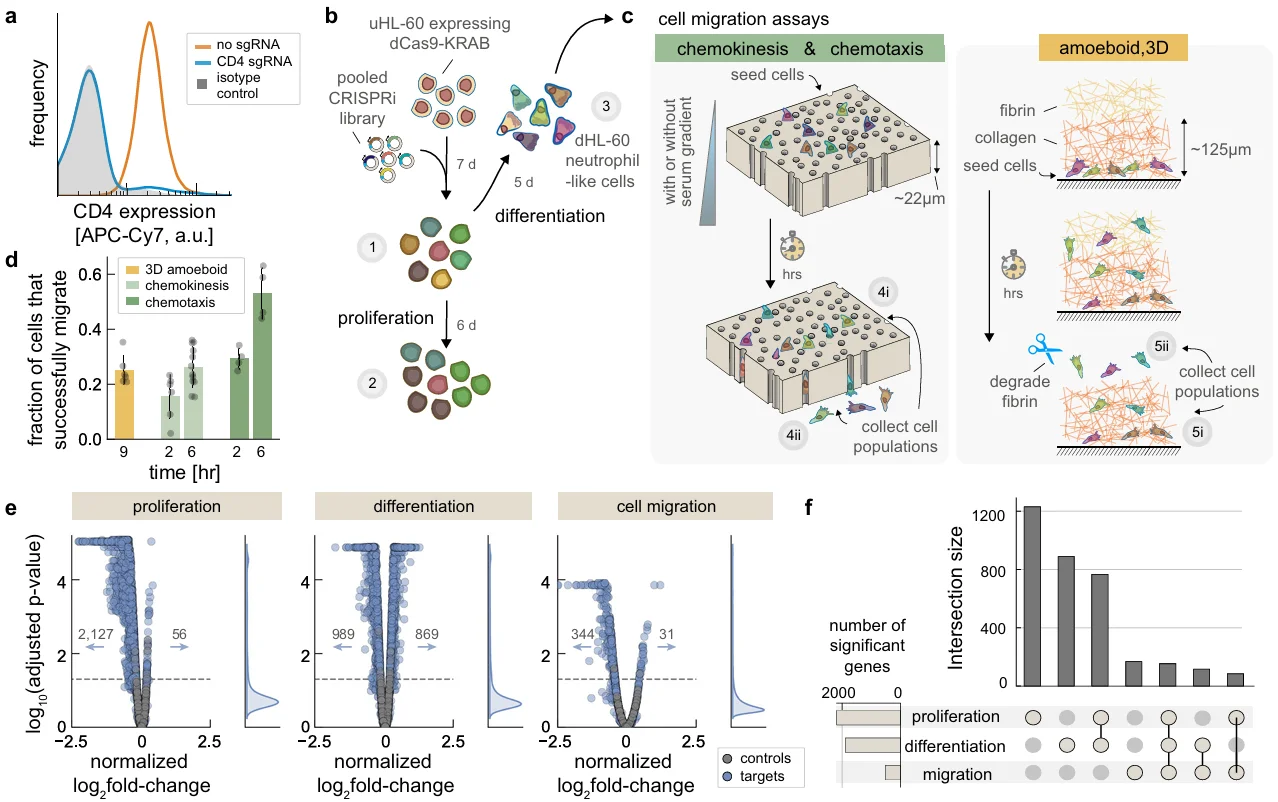
View Picture
CRISPR Library In Vitro Screening System: Viral infection (selective pressure) + cell survival/proliferation (enrichment method)
6. Replication competent HIV-guided CRISPR screen identifies antiviral factors including targets of the accessory protein Nef
Application: Identification of cellular antiviral targets
The authors generated over 1,500 replication-competent HIV-1 viruses expressing sgRNAs targeting more than 500 genes to screen for sgRNAs that enhance HIV-1 replication fitness. By serial passaging and NGS analysis in Cas9-expressing CD4⁺ T cells, several antiviral factors—including GRN, CIITA, EHMT2, and others—were identified as restricting the HIV-1 replication cycle.
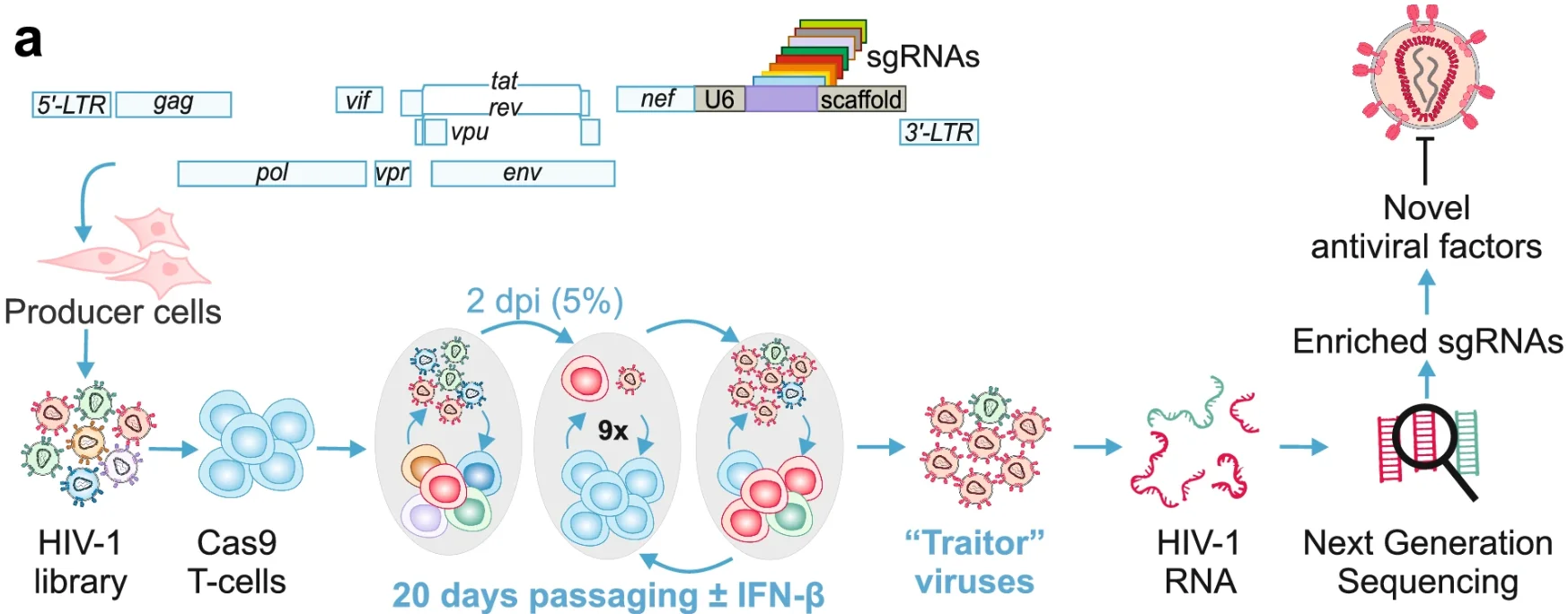
View Picture
CRISPR Library In Vitro Screening System: Tumor/immune cell co-culture (selective pressure) + cell survival/proliferation (enrichment method)
7. Integrating genome-wide CRISPR immune screen with multi-omic clinical data reveals distinct classes of tumor intrinsic immune regulators, J Immunother Cancer.
Application: Identification of immune resistance regulators
In this study, the authors applied a selective pressure system by co-culturing mouse colon cancer cells (MC38) with PmelT cells to identify unknown genes that play key regulatory roles in immunotherapy efficacy. During functional screening, for the T cell treatment group, PmelT cells were added at effector-to-target (E:T) ratios of 0.3:1 and 1:1 and cultured for 16 hours. For the non-T cell control group, an equivalent volume of T cell growth medium was added to assess in vitro sensitivity or resistance to T cell-mediated cytotoxicity. The study identified two distinct immune resistance regulators and demonstrated their potential as therapeutic targets to enhance immunotherapy efficacy. Among them, PRMT1 and RIPK1 were identified as a dual immune resistance regulator and a cytotoxicity resistance regulator, respectively. While the extent of effect varied among different types of immunotherapies, targeting PRMT1 and RIPK1 sensitized tumors to T cell-mediated killing and anti-PD-1/OX40 therapy.
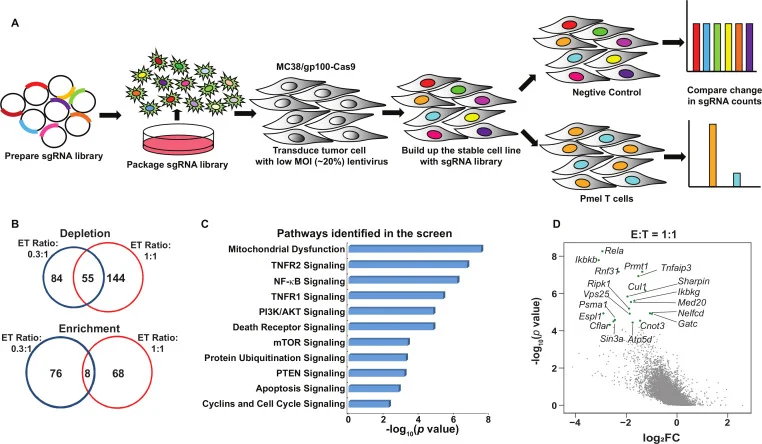
View Picture
Cytokine stimulation (selective pressure) + cell survival/proliferation (enrichment method); Cytokine stimulation (selective pressure) + flow cytometry-based enrichment (enrichment method)
8. CRISPR-Cas9 screens reveal regulators of ageing in neural stem cells, Nature.
Application: Identification of key regulatory genes involved in the activation of aged neural stem cells at the in vitro level
In this study, the authors performed CRISPR screening using a whole-genome knockout library on primary neural stem cells isolated from young and aged mice. Following viral infection of quiescent neural stem cells, specific cytokines were added to induce the transition from a quiescent to an activated state, enhancing neural stem cell activity and proliferation.Two enrichment strategies were used: Flow Cytometry-Based Enrichment: On day 4 post-activation, Ki67⁺ cells were sorted to assess sgRNA abundance, and Proliferation-Based Enrichment: On day 14 post-activation, cells were collected based on the proliferative advantage of activated neural stem cells, and sgRNA abundance was assessed.Using these complementary functional screening systems, the authors successfully identified 301 genes whose knockout specifically promoted the activation of aged neural stem cells in vitro.
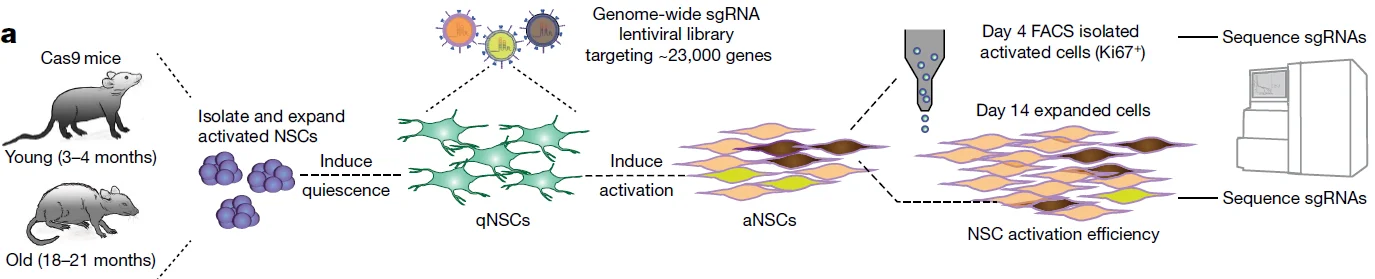
View Picture
In vivo native environment growth (selective pressure) + behavioral phenotype-based enrichment (enrichment method)
9. CRISPR-Cas9 screens reveal regulators of ageing in neural stem cells, Nature.
Application: Identification of key regulatory genes involved in the activation of aged neural stem cells at the in vivo level
To validate the functional targets identified in vitro that promote NSC activation upon knockout, the authors developed an in vivo functional screening platform in aged mouse brains. Quiescent NSCs (qNSCs) in the subventricular zone naturally activate in vivo and generate progeny that migrate to the olfactory bulb and differentiate into newborn neurons. This regenerative niche provides an ideal model for in vivo screening. The authors injected sgRNA library viruses into the lateral ventricles of aged mice to infect subventricular zone NSCs. Five weeks post-infection, genomic DNA from the olfactory bulb tissue was sequenced to analyze sgRNA abundance. Due to the limited number of cells available for in vivo screening, this platform was used for efficient validation of selected functional targets that showed significant effects in the in vitro system.
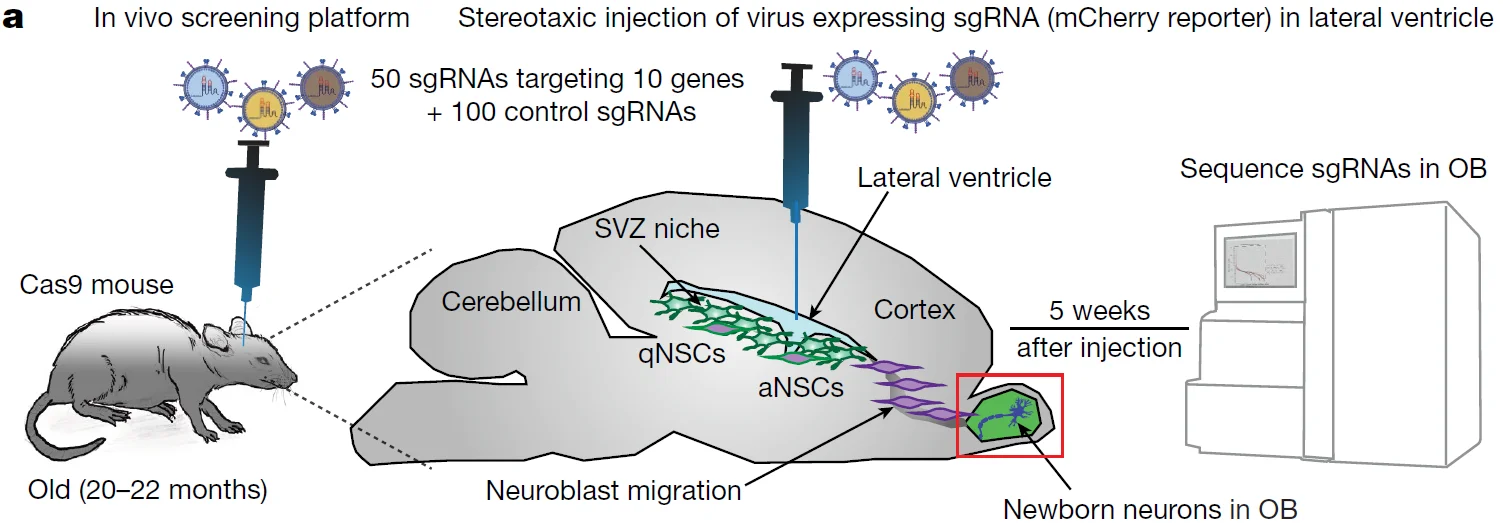
View Picture
FAQs
1. How to assess the reliability of screening results, and what controls should be included?
A CRISPR activation screen uses a catalytically - dead Cas9 (dCas9) linked to transcriptional activators(such as VP64). A gRNA library targets gene promoters or enhancers. The gRNA - dCas9 - activator complexes boost gene transcription. Cells with activated genes are selected under specific conditions. Sequencing gRNAs in these cells reveals genes related to the observed phenotype, helping study biological processes and find new therapies.
2. For flow cytometry sorting, is it sufficient that the initial cell input reaches >300* coverage, or does the sorted sample need 300* cells?
A CRISPR activation screen uses a catalytically - dead Cas9 (dCas9) linked to transcriptional activators(such as VP64). A gRNA library targets gene promoters or enhancers. The gRNA - dCas9 - activator complexes boost gene transcription. Cells with activated genes are selected under specific conditions. Sequencing gRNAs in these cells reveals genes related to the observed phenotype, helping study biological processes and find new therapies.
3. Recommended duration for drug screening, and should resistant clones be screened using positive or negative selection?
Key Differences
- Selection Pressure: Positive screens look for genes that confer resistance (survival), while negative screens look for genes that are essential (loss of viability).
- Outcome: Positive screens result in the enrichment of specific sgRNAs, whereas negative screens result in the depletion of specific sgRNAs.
- NGS Depth: Positive screens generally require lower sequencing depth compared to negative screens, which need higher depth to detect subtle
4. If the cell phenotype is not death after drug treatment, what enrichment method should be used?
- Include non-targeting sgRNAs in the library design as negative controls; these sgRNAs should not be enriched in either positive or negative selection.
- Include positive control sgRNAs (expected to be enriched in positive or negative selection). Since it is difficult to define negative outcomes in library screens, positive controls are generally more critical than negative controls.
- In experimental design, follow the controlled variable principle: the only difference between experimental and control groups should be the treatment.
5. Why perform a pre-experiment for drug screening?
- To ensure sufficient cells are collected, it is recommended to start with >500X cells before sorting; otherwise, low post-sort cell numbers may introduce experimental errors.
- The sorted sample does not need to reach 300X cells. For downstream experiments, the sorted cell number should ideally be ≥ 1 * 10⁶ cells.
6. For flow cytometry-based screening, should cells be cultured as a pool first, or can they go directly to sorting?
- Literature reports drug screens ranging from 1 day to 1 month, with most around 14 days. To achieve high enrichment of surviving cells and significant results, drug screens are generally recommended for ≥14 days.
- Whether to perform positive or negative selection depends on selective pressure:
- High selective pressure → positive selection yields more significant and reliable results
- Low selective pressure (e.g., serial passaging) → negative selection yields more significant and reliable results
7. How to best screen for drug resistance genes?
Use methods that distinguish cells based on the phenotype of interest, such as flow cytometry sorting or cell migration assays.
8. After staining pooled cells with antibodies and flow sorting for specific populations, how should samples be grouped for genomic extraction?
The pre-experiment determines the appropriate drug concentration:
- Avoid too low concentrations that have minimal killing effect
- Avoid too high concentrations that cause excessive cell death, making it difficult to collect sufficient cells for downstream NGS analysis
9. What is a CRISPR activation screen?
Pooling is recommended for ~1 week:
- To ensure enough cells for sorting, the initial input should be ≥2 × 10⁷ cells; library cells need to be expanded to obtain sufficient numbers.
- Not all sgRNAs in the library achieve high editing efficiency. Culturing pooled cells allows more cells to undergo editing, increasing the probability of gene modification.
10. What is the CRISPR drug screening?
- Use knockout library with negative selection or activation library with positive selection.
- Generally, positive selection in drug resistance screens is more reliable. For knockout libraries, positive selection identifies genes whose knockout confers resistance.
11. What is the difference between positive and negative CRISPR screens?
Flow sorting groups are typically:
- High fluorescence expression: top 5–10%
- Low fluorescence expression: bottom 5–10%