CRISPR + Flow Cytometry Help Identifying Genes Which Enable New Therapeutic Potentials for Treating Cancer


CRISPR + Flow Cytometry help identifying genes which enable new therapeutic potentials for treating cancer
CRISPR Library Screening, paired with flow cytometry, is a powerful tool for identifying gene(s) of interest. Today we set eyes on a paper published on Theranostics, Targeting tumour cell-to-macrophage communication by blocking Vtn-C1qbp interaction inhibits tumour progressino via enhancing macrophage phagocytosis. In this study, CRISPR lentiviral libraries were transduced, respectively, into tumour cell and macrophage models, and then co-cultured before being sent to Fluoresecent Activated Cell Sorting (FACS), to compare the phagocytic prowess between different KO macrophages. Subsequent experimental work was done to lock the Vtn-C1qbp interaction down as their pair of GOIs, which was shown to help tumour cells evade macrophage clearance, and thereby tumour progression.
Background
Tumour cells can evade the elimination by macrophage by overexpressing the anti-phagocytic cell surfase proteins of “don’t eat me” signals, and monoclonal antibodies targeting these “don’t eat me” signals have shown exciting potential of slowing cancer progression down. However, other unknown anti-phagocytic signals may also exist. To identify the phagocytic regulatory ligand-receptor pairs present in tumor cells and macrophages, this study utilized in vitro FACS-based phagocytic assays to directly measure phagocytic ability. Macrophages that engulfed tumor cells were screened through flow cytometry, and genes that hindered macrophage phagocytosis were plotted from a cell interaction database, leading to the identification of the Vtn-C1qbp ligand-receptor pair. This finding provides new molecular targets for targeting communication between tumor cells and macrophages, with the potential to yield new strategies for cancer therapy.
Below is an introduction to the screening process using a CRISPR library in this study.
Raw264.7 Macrophage screening
1. Transduction: Raw 264.7 cells were transduced with CRISPR sgRNA library, at a multiplicity of infection (MOI) at 0.5, to ensure only 1 sgRNA would enter each cell
2. Co-culturing and Sorting: Library-transduced RAW 264.7 cell pools were then co-cultured with GFP+ 4T1 cells for 2 hours. CD45+GFP+ phagocytic cells (those that engulfed tumor cells) were sorted and collected using flow cytometry.
3. Recovery and Identification: sgRNA sequences within the genomic DNA of sorted cells were recovered by PCR amplification and identified by high-throughput sequencing. The relative enrichment of sgRNAs in phagocytotic macrophages was calculated by the MAGeCK algorithm.
4T1 Tumour cell screening
1. Transduction: A similar CRISPR sgRNA library to what was applied to the RAW 264.7 cells were transduced into the 4T1 tumour cells.
2. Co-culturing and Sorting: Library-transduced 4T1 cell pools were co-cultured with macrophages, for CD45+GFP+Macrophages to be sorted into the phagocytic group.
3. Recovery and Identification: Similar procedure was carried out to recover the sgRNA in the sorted cells, and later sequenced to identify the relative enrichment of sgRNAs.
Experimental Results
1. Identification of Regulatory Factors of Phagocytosis in Macrophages and Tumor Cells
To systematically search for regulatory ligand-receptor interactions that modulate macrophage phagocytosis of tumor cells, the mouse genome-wide lentiviral CRISPR sgRNA library (lentiCRISPRv2-Brie) was used to transduce both macrophages and tumor cells for loss-of-function screening. A mouse macrophage cell line, Raw264.7, was chosen for its similarity to normal macrophages, exhibiting phagocytic activity and anti-tumor activity. Additionally, 4T1 cells, which can evade macrophage phagocytosis through a CD47-independent mechanism in clinical trials concerning triple-negative breast cancer (TNBC), were chosen as the tumor cell model. To visualize macrophage phagocytosis, 4T1 cells were transduced with GFP (green fluorescent protein), allowing detection of macrophage phagocytic activity using a fluorescence-activated cell sorting (FACS) machine.
Using the lentiCRISPRv2-Brie library virus with an MOI of 0.5, Raw264.7 cells were transduced to ensure that each cell was only transduced with one sgRNA. The sgRNA-transfected Raw264.7 cells were co-cultured with 4T1 cells (1:2) for in vitro phagocytic assays. After co-culturing for two hours, CD45+GFP+ phagocytes were sorted and collected. The sgRNA sequence fragments in the genomic DNA of sorted cells were recovered by PCR and identified through high-throughput sequencing. The MAGeCK (Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout) algorithm was used to calculate the relative enrichment of sgRNAs in phagocytic macrophages. A similar screening process was conducted for the 4T1 cells to identify genes regulating macrophage phagocytosis in cancer cells. Using a threshold of log2 (fold change) >0.5 and RRA score <0.25, a total of 660 potential phagocytosis-inhibiting genes were identified. The phagocytosis-inhibiting genes identified in macrophages showed strong enrichment for pathways related to phagocytosis, such as metabolism, actin cytoskeleton regulation, endocytosis, and lysosomal pathways.
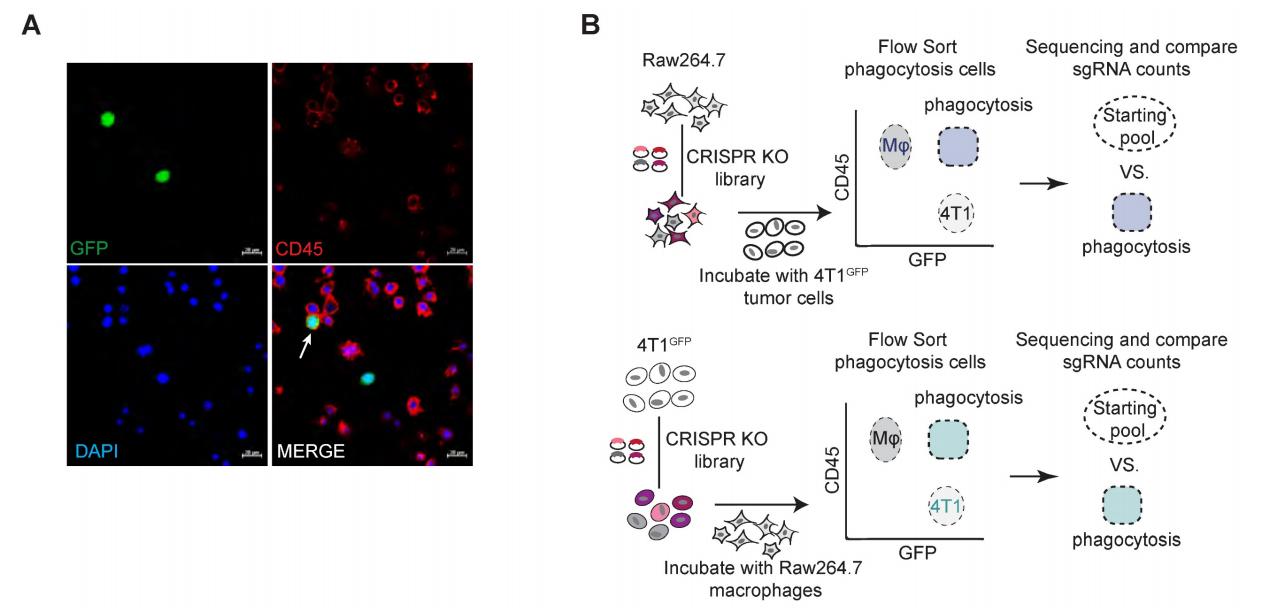
Figure 1: Genome-wide CRISPR screening identifying regulatory factors of phagocytosis.
2. Identification of the Vtn-C1qbp Ligand-Receptor Pair
Of the 660 hit genes identified in macrophages, 169 encoded cell surface proteins. Among the 1138 hit genes in tumor cells, 549 genes were considered as candidates for ligands, which showed increased expression in breast cancer tissues compared to normal tissues in the TCGA database. Candidate genes from both macrophages and tumor cells were mapped to the cell-cell interaction database from the Bader Lab at the University of Toronto, further narrowing down the candidates through flow cytometry and RNA sequencing analysis, ultimately identifying Vtn-C1qbp as a regulatory factor in the phagocytosis of tumor cells by macrophages.
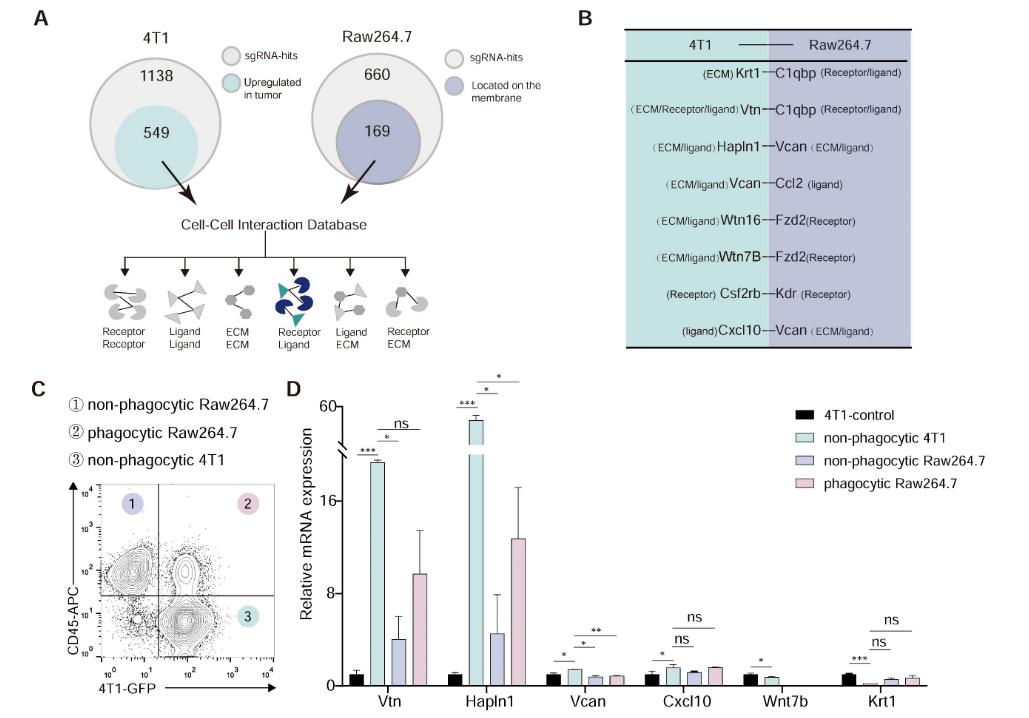
Figure 2: Vtn-C1qbp ligand-receptor pair as hypothesized modifiers of phagocytosis.
3. Validation of Vtn-C1qbp Interaction
Various experiments validated the interaction between Vtn and C1qbp. FACS analysis revealed that the membrane-bound form of C1qbp is mainly present in macrophages. Co-culture of RAW264.7 and 4T1 cells increased the proportion of C1qbp⁺ and Vtn⁺ macrophages. IP combined with western blotting analysis conducted in RAW264.7 cells showed that Vtn co-immunoprecipitates with C1qbp, primarily interacting with the αA domain of C1qbp.
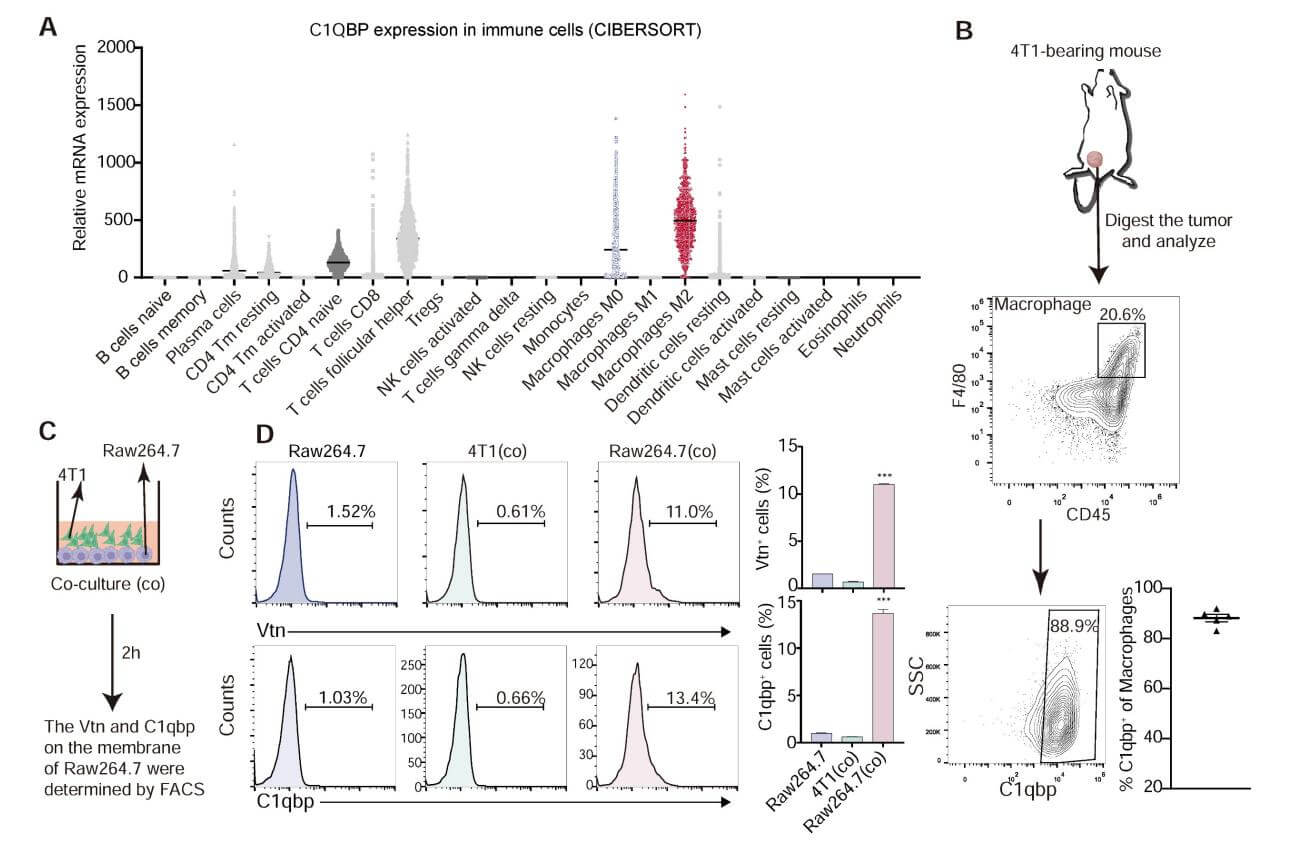
Figure 3: Vtn binds to C1qbp on the macrophage plasma membrane.
4. Impact of the Vtn-C1qbp Axis on Macrophage Phagocytosis
Knocking down Vtn in 4T1 cells enhanced macrophage phagocytic action. In vivo experiments showed that Vtn knockout tumors exhibited increased levels of phagocytosis through infiltrating tumor-associated macrophages (TAMs). Further studies indicated that the Vtn-C1qbp interaction mediates CD16-dependent recruitment of SHP1 in macrophages. Through IP-coupled mass spectrometry analysis, 296 specific binding proteins of C1qbp were identified, and KEGG pathway enrichment analysis showed that they were primarily enriched in Fcγ receptor-mediated phagocytosis and phagocytosis-related endocytosis pathways.
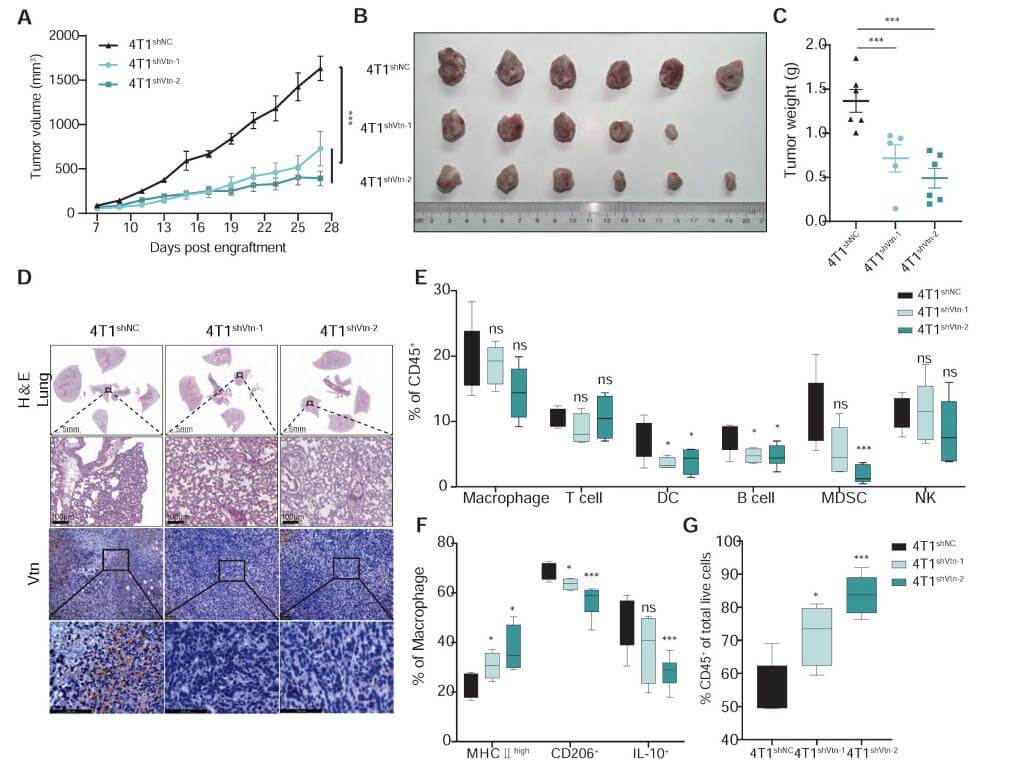
Figure 4: Vtn-C1qbp promotes differentiation of macrophages toward an M2-like phenotype.
The take-home message
This study reveals that the interaction between Vtn and C1qbp promotes tumor growth by inhibiting macrophage phagocytic ability. The findings indicate that the Vtn-C1qbp axis reduces Syk phosphorylation and recruits SHP1 mediated by FcγRIIIA/CD16, thus inhibiting macrophage phagocytosis and promoting their polarization toward an M2-like phenotype, which enhances tumor growth. Furthermore, it was found that combining the knockout of Vtn with anti-CD47 antibodies effectively enhances the phagocytic and infiltrative capacities of macrophages, reducing tumor growth in vivo. This discovery provides new molecular targets for the treatment of triple-negative breast cancer and may synergize with existing immunotherapy strategies.
Ubigene's Insights
In this study, inhibitors targeting Vtn-C1qbp were obtained through CRISPR screening, and the experiment design exploited the technique of flow cytometry to show therapeutic potential of enhancing phagocytotic capability of macrophages, and might significantly avoid tumour cells from escaping immuno-clearance and thereby holding down cancer progression. Below are some of the advantages of using FACS in a CRISPR Screening,
1. High-Throughput Analysis and Sorting: Flow sorting technology can rapidly analyze and sort thousands of cells at speeds up to tens of thousands of cells per second.
2. Multiparametric Detection: It can simultaneously measure multiple parameters of single cells, such as cell density, size, internal complexity, and other fluorescent signals.
3. High Purity and Recovery Rate: Flow sorting technology can select target cells with high purity and throughput, ensuring that the proportion of discarded sample cells is less than 5%, thus guaranteeing a high recovery rate of target cells from the samples.
4. Wide Applicability: Applicable for analysis of various cell types and biological samples, including suspension cells and appropriately treated adherent cells.
5. Flexibility: Capable of conducting positive and negative selections, as well as multiplex sorting, adapting to different experimental needs.
Ubigene can provide CRISPR screen+Flow cytometry sorting services, promotional price starting from US$8K and a target screening time fast as 8 weeks only! Feel free to contact us directly!
Reference:
Zhang C, Liu Y, Jiang J, Chen C, Duan Z, Su H, Wang S, Tian B, Shi Y, Xiang R, Luo Y. Targeting tumour cell-to-macrophage communication by blocking Vtn-C1qbp interaction inhibits tumour progression via enhancing macrophage phagocytosis. Theranostics. 2024 Apr 22;14(7):2757-2776. doi: 10.7150/thno.94537. PMID: 38773982; PMCID: PMC11103506.


