Principles and Applications of Reporter Gene Knock-in Cell Line Construction: From Mechanism to Practice


Principles and Applications of Reporter Gene Knock-in Cell Line Construction: From Mechanism to Practice

Overview of the CRISPR-Cas9 System
The CRISPR-Cas9 system, representing the third generation of gene-editing technology, has become indispensable in both basic research and clinical translation due to its high targeting specificity, programmability, and operational simplicity. Originating from prokaryotic adaptive immunity, its core components include the Cas9 nuclease and guide RNA (gRNA). Guided by gRNA, Cas9 recognizes and cleaves target DNA sequences, generating double-strand breaks (DSBs). Cells then repair these breaks through one of two endogenous pathways:
· Non-Homologous End Joining (NHEJ): A rapid but error-prone mechanism that often introduces small insertions or deletions, leading to gene knockout (KO). Ideal for disrupting gene function.
· Homology-Directed Repair (HDR): When a donor template (containing homologous arms and the desired insert) is provided, HDR enables precise integration of exogenous sequences, facilitating knock-in (KI) model construction.
Leveraging CRISPR-Cas9, researchers can efficiently generate reporter gene knock-in cell lines. These models exemplify CRISPR's utility in cellular engineering, offering advantages such as endogenous expression visualization, dynamic tracking, and high-throughput screening. As such, they have become pivotal tools in molecular mechanism studies, drug development, and functional genomics. This article systematically reviews the principles, strategies, and applications of reporter gene knock-in cell lines, providing cutting-edge insights for researchers.
Knock-in (KI) Technology: Concept and Advantages
knock-in involves inserting a target sequence (e.g., a fluorescent reporter gene) into a specific genomic locus, enabling stable expression under endogenous promoter regulation. Compared to conventional plasmid overexpression systems, knock-in cell lines offer critical benefits:
· Physiologically relevant expression levels
· Dynamic, quantitative, and spatial analysis capabilities
· Elimination of promoter leakage and random integration artifacts
CRISPR-mediated knock-in has revolutionized editing efficiency and stability, advancing precision and scalability in functional studies.
Reporter Genes and Reporter Cell Lines
1. Reporter Genes: Definition and Examples
Reporter genes are exogenous sequences whose detectable products reflect the transcriptional or translational status of target genes. They are widely used to monitor gene expression, protein localization, or signaling pathway activity.
Common types include:
| Category | Examples | Applications |
|---|---|---|
| Fluorescent proteins | GFP, mCherry, mClover3 | Real-time visualization, subcellular tracking |
| Luminescent proteins | Firefly luciferase, Renilla | High-sensitivity quantification |
| Epitope tags | FLAG, HA, His | Antibody-based detection, protein purification |
2. Reporter Cell Lines: Definition and Utility
Reporter cell lines stably express reporter genes integrated into target loci via knock-in. Their expression is governed by endogenous promoters, faithfully mirroring native gene spatiotemporal patterns. These models are indispensable for functional studies, mechanistic exploration, and drug screening.
How to Construct Reporter Knock-in Cell Lines?
To achieve high-fidelity gene expression and functional recapitulation, precise insertion strategies are essential. Current mainstream approaches include:
1. Promoter-Proximal Tagging
Insertion of the reporter gene near the target gene's transcription start site or within its first intron, preserving regulatory elements.
Advantage: Retains endogenous expression dynamics; ideal for transcriptional activity analysis.
2. In-Frame Fusion
Fusion of the reporter gene to the 5'or 3'end of the target gene, generating a functional fusion protein.
Advantage: Enables subcellular localization and trafficking studies.
3. IRES/2A Co-Expression Systems
Use of internal ribosome entry sites (IRES) or self-cleaving 2A peptides to co-express the target protein and reporter while maintaining separate translation products.
Advantage: Preserves target protein structure; suitable for functional studies.
4. Knock-in Replacement
Complete replacement of the endogenous coding sequence with the reporter gene, driven by the native promoter.
Advantage: Useful for gene function ablation or substitution.
Ubigene’s CRISPR-U™ Platform employs high-throughput gRNA optimization and donor vector design to enhance knock-in efficiency and specificity, achieving 5–10× higher success rates than conventional methods across 300+ mammalian cell lines. Contact us now>>>
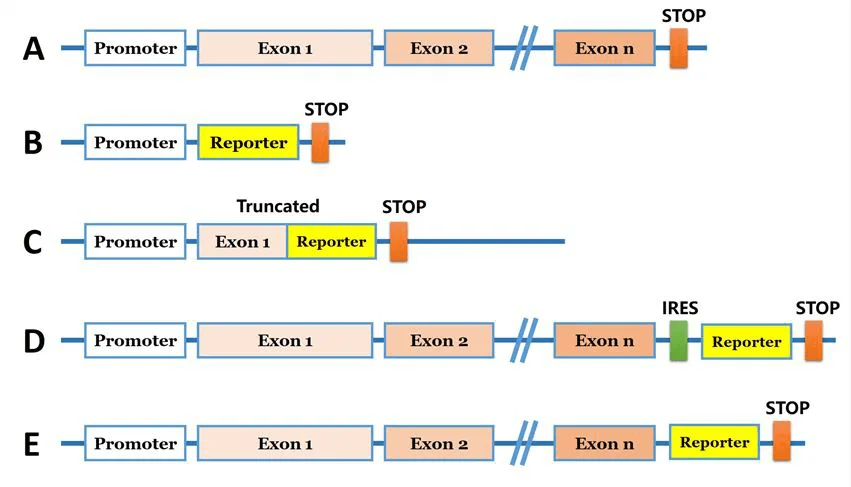
Figure 1: Common strategies for reporter gene knock-in
Comprehensive Analysis of Reporter Knock-in Cells Applications
1. Monitoring Endogenous Promoter Activity and Studying Gene Regulatory Mechanisms
Reporter gene knock-in technology enables monitoring of specific promoter activity within native chromatin contexts, overcoming the interference caused by traditional plasmid-based systems. For example, integrating a reporter gene (e.g., EGFP or luciferase) downstream of a target gene's transcription start site allows real-time tracking of promoter activity. This is particularly valuable for studying transcription factor regulation, epigenetic modifications, and non-coding regulatory elements.
Case Study:
In research on the SREBP1 gene, scientists used CRISPR/Cas9 to knock in a luciferase gene at the SREBP1 locus, generating a HEK293-SREBP1-T2A-luciferase reporter cell line. One allele precisely integrated luciferase, while the other allele was functionally knocked out via a 14-bp deletion. Luciferase activity in this cell line closely correlated with endogenous SREBP1 promoter activity, making it a powerful tool for dynamic transcriptional monitoring.
Advantages:
1. Accurately reflects endogenous gene regulation
2. Facilitates promoter mutation analysis, transcription factor screening, and signaling pathway activity assays
3. Enables kinetic studies of promoter responses to drugs or external stimuli
2. Protein Expression, Localization, and Subcellular Trafficking Dynamics
Fusing fluorescent tags (e.g., GFP, mCherry) to the N- or C-terminus of target proteins is a classic strategy for functional studies. CRISPR/Cas9-mediated seamless integration of tag genes into endogenous loci produces fusion proteins that retain native expression regulation while enabling real-time imaging, quantitative expression analysis, and organelle localization studies.
Case Study:
In the GFP-LC3 reporter cell model, researchers knocked in green fluorescent protein (GFP) at the N-terminus of the endogenous MAP1LC3B locus in 293FT cells. The resulting GFP-LC3 fusion protein, driven by the native promoter, is widely used in autophagy research. This system distinguishes free GFP from LC3-labeled autophagosomes, showing clear signal differences under rapamycin induction or chloroquine (CQ) inhibition, enabling dynamic assessment and quantitative analysis of autophagy levels.
Advantages:
1. Eliminates the need for antibodies in protein detection
2. Ideal for studying translation, post-translational modifications, complex formation, and degradation
3. Supports high-content imaging and live-cell tracking
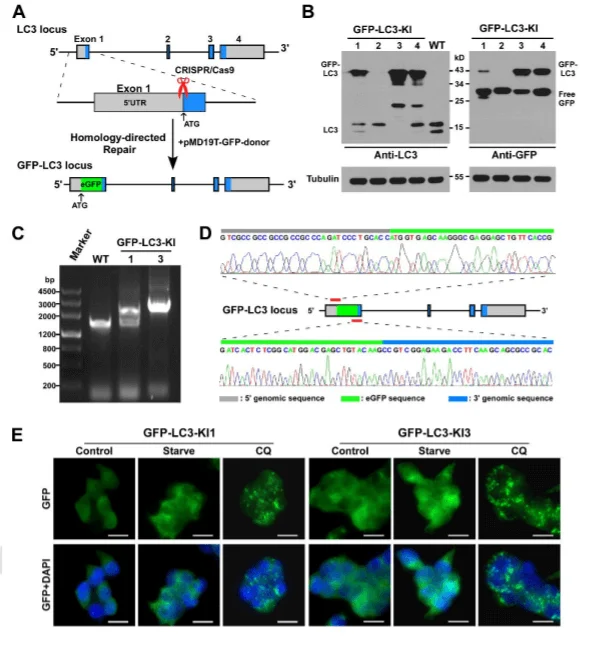
Figure 2: Knock-in of GFP-LC3 at the MAP1LC3B locus in 293FT cells
3. Target identification and evaluation of drug candidates
Reporter gene knock-in cell lines serve as high-sensitivity, low-background platforms for drug screening. By fusing reporter genes (e.g., luciferase or fluorescent proteins) to drug target genes, researchers can monitor changes in reporter signals to assess target gene expression or functional states, accelerating the identification of pathway-modulating compounds.
Case Study:
A triple-labeled HeLa cell line (nuclear protein Histone 1-mTagBFP2, cytoskeletal β-Tubulin-mClover3, and autophagy receptor SQSTM1-mRuby3) was used to screen kinase inhibitors' effects on autophagy via fluorescence colocalization and live-cell imaging. Both PLK1 inhibitor BI-6727 and PIM kinase inhibitor CX-6258 induced significant accumulation of autophagic vesicles, mimicking the effects of the known autophagy blocker hydroxychloroquine. This validated the system's utility in pharmacodynamic studies.
Advantages:
1. Predicts drug sensitivity and elucidates mechanisms
2. Enables high-throughput, automated primary screening
3. Reduces false positives caused by expression system artifacts
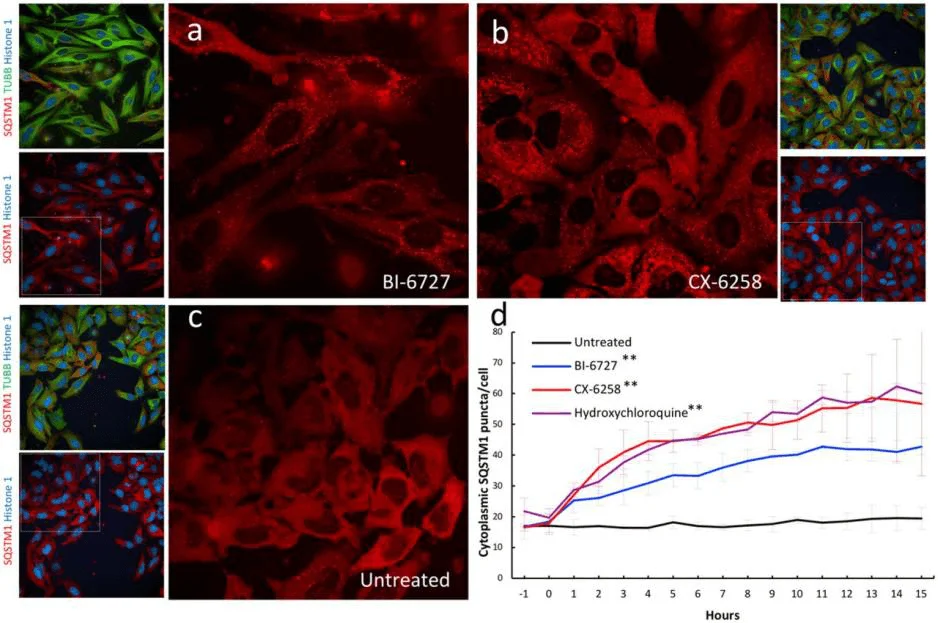
Figure 3: Validation of kinase inhibitors that induce autophagic vesicle accumulation in triple-labeled HELA cell lines
Conclusion and Future Perspectives
The widespread adoption of reporter gene knock-in cell lines marks a new era in molecular and cellular biology, characterized by precision visualization, dynamic quantification, and high-throughput capabilities. These models not only deepen the understanding of gene function but also provide robust technical support for drug development, synthetic biology, and personalized medicine.
As CRISPR tools evolve—integrated with single-cell sequencing, AI-assisted drug screening, and 3D organoid technologies—reporter cell lines will transition into advanced research platforms, bridging the gap between basic research and clinical translation.
Ubigene offers customized reporter gene knock-in cell line construction services covering diverse tag types and integration strategies. High-purity monoclonal cell lines can be delivered in as little as 6 weeks. Contact our technical experts for tailored solutions!
References
[1] Establishment of a HEK293 cell line by CRISPR/Cas9- mediated luciferase knock-in to study transcriptional regulation of the human SREBP1 gene. Biotechnol Lett, 2018, (11-12):1495-1506.
[2] CRISPR/Cas9 mediated GFP knock-in at the MAP1LC3B locus in 293FT cells is better for bona fide monitoring cellular autophagy. Biotechnol J, 2018, 13(11): e1700674.
[3] 3.A Versatile Vector System for the Fast Generation of Knock-in Cell Lines with CRISPR, 2020, doi: https://doi.org/10.1101/2020.02.06.927384


