Smaller Scale, Broader Applications: Cloning Optimization Makes CRISPR Library Screens More Compact


Smaller Scale, Broader Applications: Cloning Optimization Makes CRISPR Library Screens More Compact
In CRISPR screening experiments, the uniformity of sgRNA libraries is a key determinant of data reliability. However, conventional library construction strategies often suffer from distribution skew, resulting in certain sgRNAs being underrepresented or even lost. To mitigate false negatives, researchers are typically required to perform screens at very high cell coverage (≥300×). This requirement poses major challenges for cell types that are difficult to expand, such as iPSC-derived cells or primary cells, thereby limiting the broader application of CRISPR screens.
Recently, Chow ED and colleagues reported in Genome Biology (Heo SJ et al., 2024) an improved sgRNA library cloning strategy that combines bidirectional template design with optimized cloning conditions. This approach enables the construction of a more compact and uniform CRISPR library (LGR library), significantly reducing library skew ratio. As a result, robust genome-wide screening performance can be achieved even at substantially lower cell coverage levels.
Research background: from cloning bias to screening bottlenecks
CRISPR screening relies on the even representation of sgRNAs across a cell population. However, conventional library construction introduces cloning and amplification biases, causing low-abundance sgRNAs to be underrepresented and easily lost during screening. To compensate, researchers are often forced to increase cell coverage, which substantially raises both cost and technical barriers. Although some efforts have explored compressed library designs—such as dual-sgRNA systems—to reduce scale, these approaches introduce risks of recombination and statistical complexity. In contrast, Chow and colleagues focused on optimizing the cloning process itself, offering a simpler and more effective solution to this fundamental problem.

Optimization strategy: toward higher-quality CRISPR libraries
Through stepwise experimental refinements, the team established a generalizable workflow that yields more uniform sgRNA representation across genome-scale libraries.
1. Template synthesis: bidirectional sgRNA library design
While many researchers routinely order oligo pools containing both sense and antisense sgRNA sequences, prior studies rarely emphasized this practice or provided supporting evidence for its utility. Chow and colleagues were the first to explicitly demonstrate that using bidirectional sgRNA templates during cloning reduces library bias and guide loss, compared to traditional unidirectional designs (Fig. 1E, F).
2. Template cloning: minimizing amplification cycles
The authors compared three commercial DNA polymerases for PCR-based cloning and selected the one with optimal performance (Fig. 1A, B). By testing libraries subjected to either 1 or 15 PCR cycles, they showed that excessive amplification introduces subtle distribution skew (Supplementary Fig. S4) and promotes over-representation of non-target fragments. Careful adjustment of PCR cycle number and template input effectively minimized these artifacts, improving library fidelity.
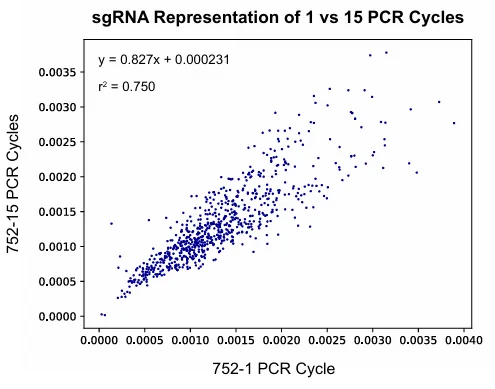
Supplementary Fig. S4. Additional PCR cycles introduce subtle biases in sgRNA library representation.
3. Fragment insertion: low-temperature elution
In a small library containing 192 sgRNAs, elution at 37°C markedly improved sgRNA uniformity compared to high-temperature elution at 70°C (Fig. 1A, B). However, when scaling up to a 752-sgRNA library, elution at 37°C was insufficient to overcome library bias arising from intrinsic sgRNA melting temperature (Tm) differences (Fig. 1C, D). By combining this approach with the previously optimized conditions and further lowering the elution temperature to 4°C, the team achieved a genome-wide library in which sgRNA Tm no longer correlated significantly with guide abundance (Fig. 1G).
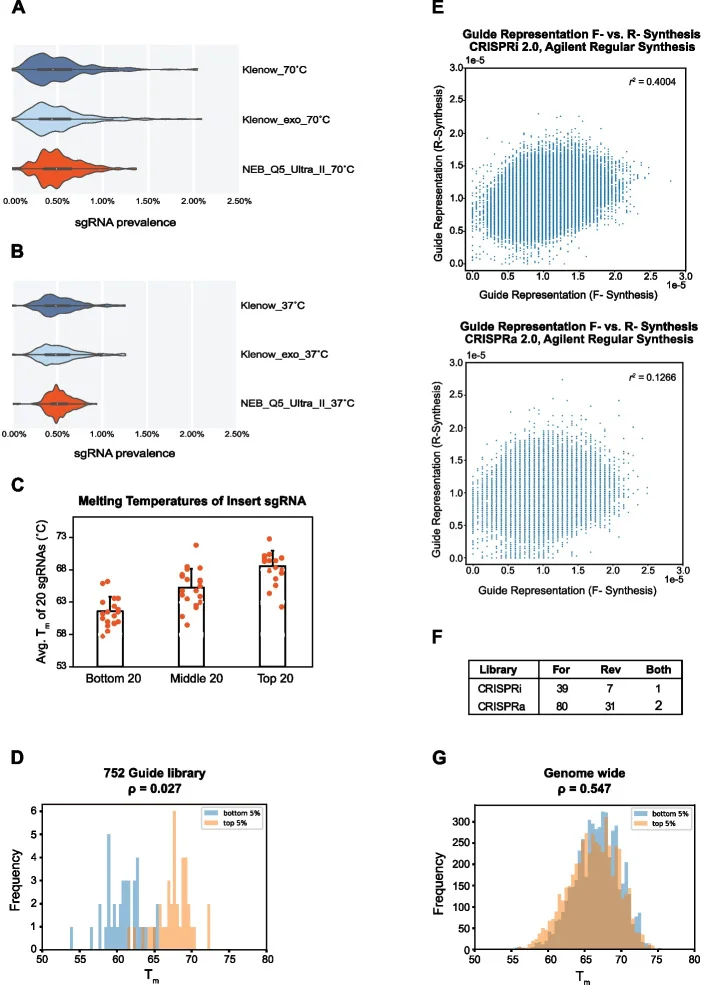
Figure 1. Factors affecting sgRNA uniformity in CRISPR libraries
4. CRISPR Library cloning: single-step, whole-genome approach
Conventional CRISPRi/a v2 libraries are divided into seven sub-libraries that are cloned independently, with each sub-library exhibiting distinct skew. Consequently, the overall library distribution is influenced by sub-libraries with high variance. Chow and colleagues applied their optimized conditions to clone the entire genome-wide library in a single step. The resulting “LGR” library (optimized cloning) demonstrated more uniform sgRNA representation and reduced guide loss compared to the “Legacy” library (conventionally cloned) (Fig. 2).
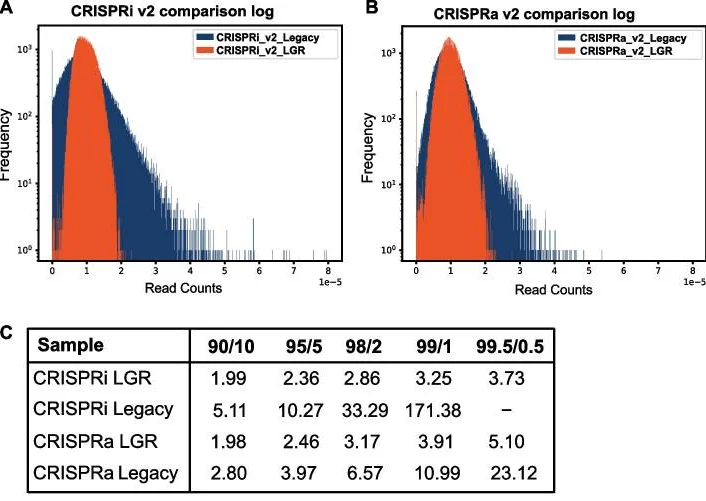
Figure 2. Optimized cloning strategy improves genome-wide CRISPR library uniformity
Performance demonstration: LGR library enables superior screening
1. LGR library allows lower cell coverage
To evaluate whether the LGR library could support screens at reduced cell coverage, Chow and colleagues reproduced a previous genome-wide screen by Horlbeck MA et al. using the LGR library (Fig. 3A). Comparison of essential gene hits showed that at 1000× coverage, the number of essential genes identified was comparable to the original study, with 1,366 essential genes overlapping (Fig. 3B, D). Notably, the area under the curve (AUC) at 1000× coverage reached 0.937 (Fig. 3C), demonstrating high precision in essential gene identification. Remarkably, at only 100× coverage, the LGR library still achieved an AUC of 0.949, with greater overlap in hits and similar phenotype scores compared to the 1000× coverage screen (Fig. 3C–E).
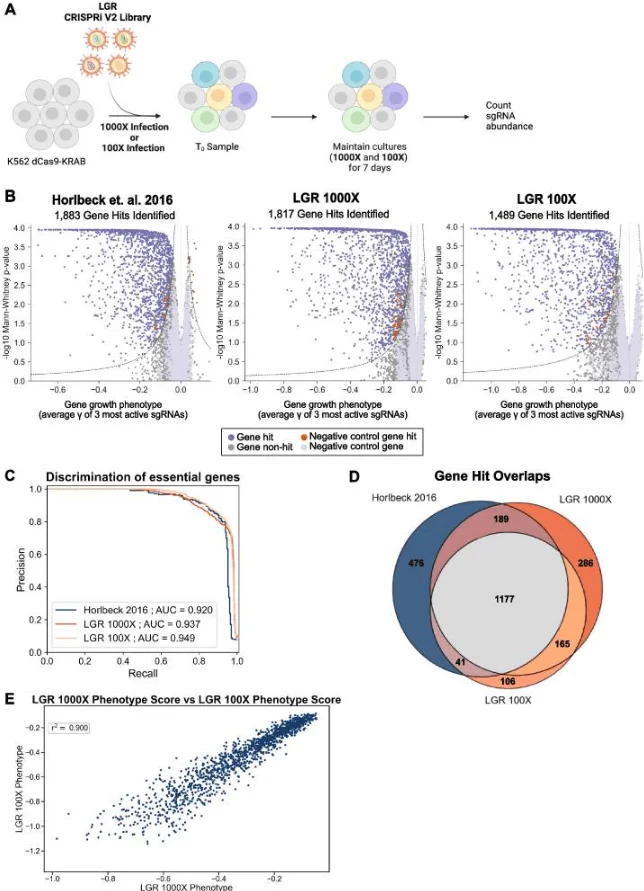
Figure 3. The optimized LGR library maintains performance comparable to existing libraries at reduced cell coverage
A major challenge in screening at reduced cell coverage is sgRNA dropout. The LGR library exhibits nearly consistent library skew ratios and minimal sgRNA loss even at 200×, 100×, and 50× coverage (Fig. 4), suggesting that, in principle, genome-wide screens can be performed with the LGR library at cell coverage as low as 50×.
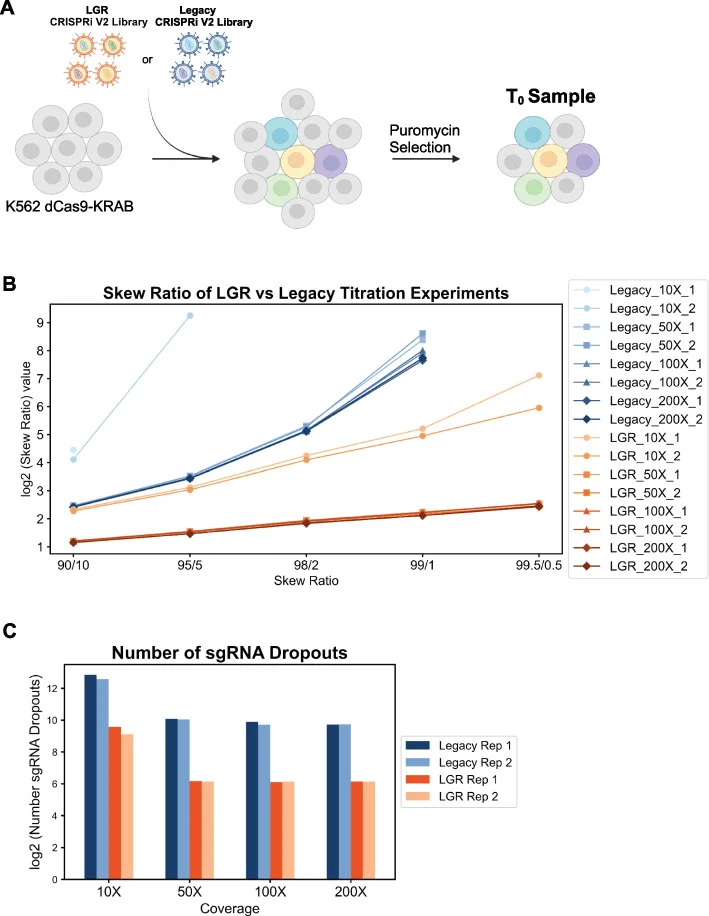
Figure 4. Comparison of sgRNA uniformity and dropout between LGR and conventional libraries post-transduction
2. LGR library yields more candidate gene hits
To assess whether the LGR library could support high-quality screens at low coverage, Chow and colleagues performed Dasatinib survival screens using both conventional CRISPRi libraries and the LGR library, comparing their ability to comprehensively identify genes associated with Dasatinib resistance and sensitivity (Fig. 5A, B). The results showed that the LGR library captured more essential gene hits while producing a comparable number of drug-related gene hits relative to the conventional library (Fig. 5C, D). Furthermore, as the false discovery rate (FDR) threshold was tightened, the LGR library continued to yield a higher number of candidate gene hits (Fig. 5E).
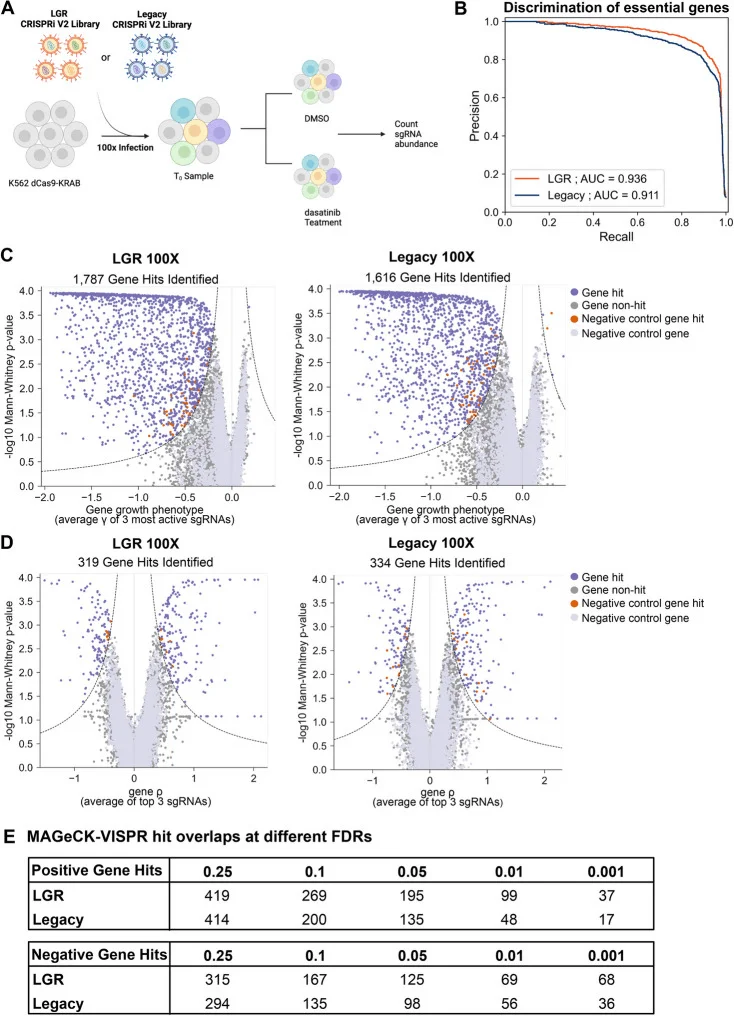
Figure 5. The LGR library identifies more candidate gene hits at 100× coverage
The mediator complex (GO ID: 0016592) and the oxidative phosphorylation pathway (KEGG ID: hsa00190) have been identified as potential drug targets synergistic with Dasatinib. Importantly, the LGR library yielded more hits among genes associated with both the mediator complex and oxidative phosphorylation pathways (Fig. 6), further demonstrating that the LGR library not only maintains high precision but also uncovers a greater number of screening hits.
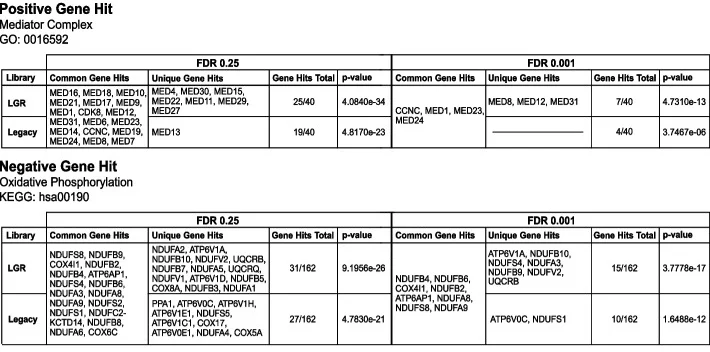
Figure 6. Gene hits identified by LGR and conventional libraries in the mediator complex (top) and oxidative phosphorylation pathway (bottom)
Technical advantages and significance
In summary, this optimized cloning strategy offers several key advantages:
1. significantly reduced cell requirements.
The improved cloning approach produces a more uniform sgRNA library, allowing genome-wide screens to be performed at cell coverage as low as 50× for a given library size. Conversely, with the same number of cells, researchers can perform larger-scale sgRNA screens.
2. More reliable screening results with reduced false negatives and positives.
The uniform sgRNA distribution minimizes loss or differential expression of guides, enhancing the accuracy and reproducibility of screening outcomes. This also enables more biological replicates under the same cell constraints, further improving data reliability.
3. expanded applicability of CRISPR library screening.
This optimized cloning method makes genome-wide CRISPR screens feasible in technically challenging cell types or screening models, such as adherent cells for FACS-based imaging, iPSC-derived cells, and primary cells. Moreover, this straightforward improvement is broadly applicable across diverse CRISPR systems, including Cas9, Cas12, and Cas13, as well as derivative technologies such as base editing and CRISPRoff.
If you are struggling with the high cell amount requirements and lengthy timelines of conventional CRISPR library screens, Ubigene now offers a compact and efficient solution. Leveraging our CRISPR-iScreen™ platform combined with in vitro/in vivo screening services and the zero-code iScreenAnlys™ CRISPR library analysis platform, you can:
- · Access Screening-ready Cell Pools (400+ available) directly, bypassing library construction and enabling immediate experiments;
- · Perform screens in both in vitro models and in vivo settings, faithfully recapitulating disease microenvironments to accurately identify key gene targets;
- · Using the iScreenAnlys™ CRISPR library analysis platform, users can import data and automatically perform quality control, differential analysis, pathway enrichment, and target ranking—without any programming skills—while generating publication-quality volcano plots, heatmaps, and other figures.
Currently, Ubigene offers a special promotion for in vitro CRISPR library screening services, with turnaround times as short as 6 weeks. Generate high-quality results using fewer cells and lower costs, giving your research a clear advantage in both speed and accuracy.
Contact us for more technical support >>
Reference
Heo SJ, Enriquez LD, Federman S, et al. Compact CRISPR genetic screens enabled by improved guide RNA library cloning. Genome Biol. 2024; 25(1): 25.
doi: 10.1186/s13059-023-03132-3.



