When CRISPR Library Screens Meet Tumor Metabolism


When CRISPR Library Screens Meet Tumor Metabolism: Why Context Is Everything
I. Background
In recent years, CRISPR-based genetic library screening has become a de facto standard technique in cancer research. Leveraging this powerful tool, researchers can systematically identify genes that are essential for the survival and proliferation of cancer cells—so-called core dependencies. Some of these screening-derived targets have even advanced into clinical trials, underscoring the transformative impact of CRISPR screening in both fundamental research and translational oncology.
However, a critical but often overlooked issue lies in the experimental context: most CRISPR library screens are performed under highly idealized, standardized cell culture conditions. In such settings, glucose levels are far higher than physiological concentrations, amino acid supplies are plentiful and complete, and oxygen availability approaches saturation.Compared with the in vivo tumor microenvironment—where nutrient deprivation, hypoxia, and metabolic stress coexist—these culture conditions are akin to a “five-star hotel,” whereas tumor cells in the body are struggling in a state closer to “survival in the wilderness.”
As a consequence, while standardized culture conditions facilitate large-scale, high-throughput screening, they fail to capture the pathological and physiological constraints of real tumors. This discrepancy may obscure gene functions that are only revealed under stress or nutrient-limited conditions. In other words, the dependencies identified from conventional CRISPR screens may reflect cancer cell survival strategies in artificially favorable environments, rather than those that operate under clinically relevant metabolic stresses.
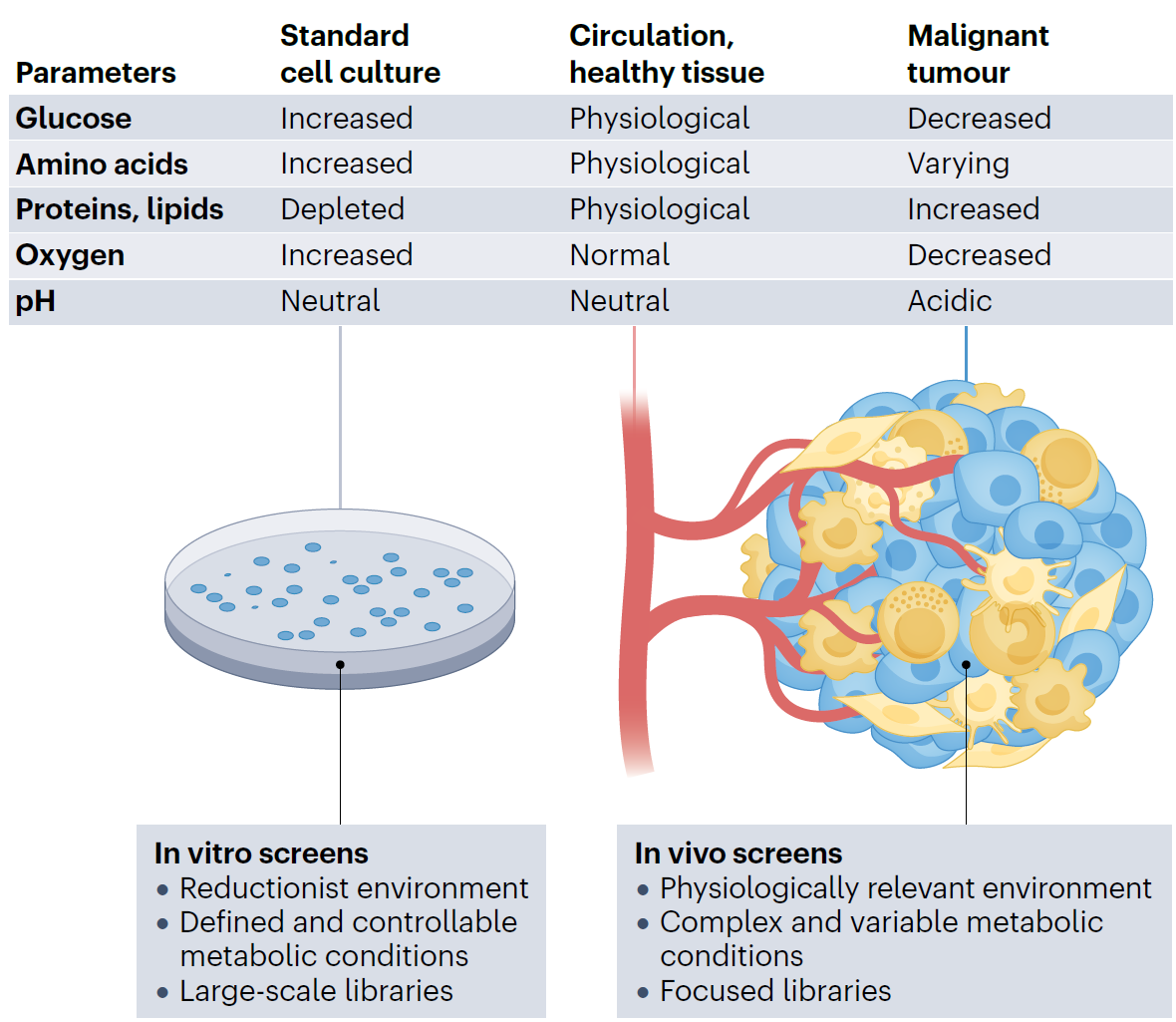
Figure 1. Metabolic Contexts in CRISPR Screening
This raises a critical question: under such non-physiological conditions, can CRISPR library screening results truly reflect what cancer cells depend on in vivo?
To address how the metabolic environment shapes screening outcomes, Johannes Zuber and Wilhelm Palm published a commentary in Nature Reviews Cancer entitled "Modelling and deciphering tumour metabolism in CRISPR screens." This article highlights the importance of contextualizing CRISPR-based screens within physiologically relevant metabolic environments, and aims to increase awareness among researchers that environmental conditions profoundly influence the interpretation of CRISPR screening results.

II. Research Content
1.How Do Metabolic Conditions Alter CRISPR Screening Outcomes?
To investigate how different culture environments affect CRISPR screen results, the authors designed a series of experimental settings that mimic distinct metabolic states of tumors. By systematically comparing findings obtained under these varied conditions, they arrived at a clear conclusion: the metabolic environment profoundly shapes the genetic dependencies of cancer cells.
The researchers established cell models under single-variable culture conditions—modulating glucose, oxygen, and amino acid availability—to delineate how individual environmental factors influence gene essentiality. The results revealed striking context-dependent differences:
● Under glucose deprivation, cancer cells were forced to rely on mitochondrial oxidative phosphorylation (OXPHOS) to sustain ATP production.
● In contrast, under high-glucose conditions, the same cells survived predominantly via glycolysis, showing little dependence on OXPHOS.
● In oxygen-limited (hypoxic) environments, OXPHOS ceased to be a critical pathway; instead, lipid metabolic pathways emerged as essential for cell growth and survival.
Beyond carbon metabolism, the researchers also explored nutrient stress driven by amino acid restriction. Under limited amino acid availability, some tumor cells switched to scavenging extracellular proteins as alternative sources of carbon and nitrogen—an adaptation that depended on lysosomal degradation and recycling. Notably, LYSET was identified as a key transporter mediating this process.To further approximate physiological conditions, the authors employed culture media formulated to reflect in vivo nutrient concentrations, revealing a subset of “hidden” dependency genes—genes that appeared non-essential under standard high-nutrient conditions but became critical in nutrient-poor environments. These context-specific vulnerabilities exhibited greater pathological and clinical relevance.
Collectively, these findings lead to a compelling conclusion: cancer cell gene dependencies are not fixed entities but are dynamically sculpted by the metabolic microenvironment. In other words, the tumor context does not merely modulate but fundamentally defines which genes are essential for survival. Ignoring the influence of metabolic conditions may therefore obscure the most crucial—and clinically actionable—targets.
2.Strategies to Bridge the Gap: Bringing the “Real World” into CRISPR Library Screens
To overcome the limitations described above, the authors propose two complementary strategies aimed at more accurately identifying cancer cell dependencies within physiologically relevant contexts.
(1) Reconstructing the Tumor Metabolic Microenvironment in vitro
The first strategy focuses on in vitro systems that faithfully recapitulate specific metabolic features of tumors—such as glucose deprivation, hypoxia, or high-protein conditions—to probe how individual environmental stresses reshape gene essentiality. For example, RNAi-based screens have shown that under glucose limitation, cancer cells shift toward a dependency on mitochondrial oxidative phosphorylation (OXPHOS). Similarly, under high-protein culture conditions, the lysosomal transporter LYSET was identified as a crucial factor enabling cancer cells to utilize extracellular proteins as alternative nutrient sources.
An alternative in vitro approach involves the use of "physiologic" culture media formulated to mimic the nutrient composition of the in vivo tumor milieu. CRISPR screens conducted under these conditions can reveal conditionally essential genes that remain undetectable under conventional, nutrient-rich culture systems—thereby exposing vulnerabilities with stronger pathological and translational relevance.
(2) Performing CRISPR Screens in vivo
The second strategy involves conducting CRISPR library screens directly in animal models, such as subcutaneous tumor xenografts in mice, where cancer cells experience the complex and dynamic metabolic landscape of living tissue. Such in vivo screens have uncovered unique dependencies—for instance, heme biosynthesis enzymes that are indispensable for tumor survival in vivo but remain undetected in standard in vitro settings.
Nevertheless, in vivo CRISPR library screening also faces substantial technical constraints. The limited number of analyzable cells often makes it infeasible to maintain genome-scale coverage, necessitating the use of focused or pathway-specific libraries. In addition, pre-screening steps involved in CRISPR library construction and transplantation may introduce biases that influence the ultimate screening outcomes.
3.The Road Ahead: Towards More Physiologically Relevant CRISPR Screening
The authors emphasize that the future of CRISPR library screening will rely on continuous technological innovation to more faithfully capture the complex metabolic reality of tumors. Several directions for improvement are proposed:
● Smaller, more efficient libraries: With advances in sgRNA design optimization, CRISPR libraries can be further miniaturized to create compact yet high-precision screening collections. This downsizing not only enhances the feasibility of in vivo screening but also facilitates multidimensional experiments under more complex conditions—such as incorporating metabolic perturbations, different cell line models, or pharmacological treatments.
● Inducible CRISPR systems: Inducible genome-editing systems allow temporal control of gene knockout, keeping the CRISPR machinery inactive during pre-transplantation culture and activating it only after cells have engrafted in vivo. Building inducible CRISPR–Cas9 platforms is thus essential to minimize artifacts caused by pre-screening stress or nutrient conditions, thereby enabling a more accurate dissection of gene function within physiologically relevant settings.
● More realistic modeling: Future screening efforts should make broader use of physiologic or plasma-like media, tumor interstitial fluid–based formulations, and advanced culture systems such as organoids or co-culture models. These platforms can better reproduce the tumor microenvironment and its metabolic heterogeneity.
● Multidimensional integration: CRISPR screens should evolve toward designed capable of capturing the multi-layered complexity of tumor metabolism, for instance, by simultaneously probing the effects of glucose limitation and hypoxia, or integrating metabolomic, transcriptomic, and phenotypic readouts.
● Complementarity between in vitro and in vivo approaches: In vitro screening offers unmatched controllability and scalability for systematic, high-throughput discovery, while in vivo screening provides pathophysiological relevance crucial for validating and translating findings. A synergistic workflow combining both settings will be key to identifying robust, clinically meaningful cancer dependencies.
4.Implications for Research and Drug Development
From a research perspective, current dependency databases—such as DepMap—are largely derived from screens conducted under standard culture conditions, which often underestimate condition-specific or context-dependent essential genes. Future studies should incorporate metabolic environment as a critical experimental variable, performing parallel CRISPR screens under diverse nutrient, oxygen, or stress conditions. Such efforts will yield a more accurate and comprehensive functional map of cancer cell dependencies, illuminating the adaptive and plastic nature of tumor metabolism.
From a drug development perspective, gene dependencies that emerge only under noncanonical or stress-induced metabolic conditions may pinpoint targets with higher clinical relevance. These context-specific vulnerabilities can guide the identification of druggable targets, deepen our understanding of the metabolic "survival limits" of cancer cells, and reveal the molecular underpinnings of therapy resistance. Ultimately, integrating environmental context into CRISPR-based discovery will support tumor subtype stratification and advance personalized precision oncology.
III. Overall Summary
The metabolic environment fundamentally determines CRISPR screening outcomes—the metabolic state of tumor cells directly shapes their genetic dependencies. Results obtained under standard, nutrient-rich culture conditions often reflect an idealized setting that fails to capture the true dependencies operating within the tumor microenvironment. By combining in vitro and in vivo screening approaches, researchers can achieve complementary strengths: in vitro screens provide high efficiency and scalability for systematic discovery, while in vivo screens offer greater physiological and pathological relevance.
Integrating both strategies is essential to construct a comprehensive and reliable map of cancer gene dependencies. Looking forward, CRISPR screening will continue to evolve toward more realistic tumor models, incorporating metabolic perturbations, organoid systems, and co-culture models to better recapitulate the complexity of the tumor microenvironment. Such efforts will enable the identification of drug targets with stronger translational and clinical potential.
For researchers, this serves as a critical reminder: before interpreting any CRISPR screening result, one must first ask—under what environmental conditions were these dependencies defined? Only by answering this question can findings be properly contextualized within their biological and clinical frameworks.
IV. CRISPR Library Screening Services
Accurate CRISPR screening outcomes rely on rigorous experimental design, and efficient execution requires a reliable partner. Ubigene now offers comprehensive in vitro + in vivo CRISPR library screening services, spanning from controlled in vitro metabolic modeling to the complex, physiologically relevant in vivo environment, ensuring that key target genes are systematically captured. Coupled with our proprietary iScreenAnlys™ CRISPR library analysis platform, screening data can be processed and interpreted with a single click, streamlining the transition from raw data to actionable insights.
We also provide customized screening solutions tailored to your specific research needs. Whether you are performing positive or negative screens, or require specialized cell culture conditions, our team can flexibly adapt to your experimental requirements.
Contact us to get technical support>>
Reference
Zuber J, Palm W. Modelling and deciphering tumour metabolism in CRISPR screens. Nat Rev Cancer. 2025 Jan;25(1):1-2. doi: 10.1038/s41568-024-00758-8. PMID: 39354071.
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
Birsoy, K. et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 508, 108–112 (2014).
Jain, I. H. et al. Genetic screen for cell fitness in high or low oxygen highlights mitochondrial and lipid metabolism. Cell 181, 716–727.e711 (2020).
Pechincha, C. et al. Lysosomal enzyme trafficking factor LYSET enables nutritional usage of extracellular proteins. Science 378, eabn5637 (2022).
Rossiter, N. J. et al. CRISPR screens in physiologic medium reveal conditionally essential genes in human cells. Cell Metab. 33, 1248–1263.e1249 (2021).
Biancur, D. E. et al. Functional genomics identifies metabolic vulnerabilities in pancreatic cancer. Cell Metab. 33, 199–210.e198 (2021).
Zhu, X. G. et al. Functional genomics in vivo reveal metabolic dependencies of pancreatic cancer cells. Cell Metab. 33, 211–221.e216 (2221).
Michlits, G. et al. Multilayered VBC score predicts sgRNAs that efficiently generate loss-of-function alleles. Nat. Methods 17, 708–716 (2020).
Sullivan, M. R. et al. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. eLife 8, e44235 (2019).
Abbott, K. L. et al. Screening in serum-derived medium reveals differential response to compounds targeting metabolism. Cell Chem. Biol. 30, 1156–1168.e1157 (2023).



