From Genetic Code to Cell Therapy: Genome-wide CRISPR Screening Reveals Novel Regulatory Mechanisms in iTreg Cell Therapy


From Genetic Code to Cell Therapy: Genome-wide CRISPR Screening Reveals Novel Regulatory Mechanisms in iTreg Cell Therapy

Regulatory T cells (Tregs),often referred to as the “guardians” of the immune system, maintain immune tolerance through the lineage-defining transcription factor FOXP3. Dysfunction of Tregs can lead to various diseases, including autoimmune disorders. However, in vitro-induced Tregs (iTregs) suffer from unstable FOXP3 expression, which has long been a major obstacle to their clinical translation in cell therapy. A 2025 study published in Nature, “Genome-wide CRISPR screen in human T cells reveals regulators of FOXP3,” employed genome-wide CRISPR screening to systematically identify FOXP3 regulatory genes. The study revealed RBPJ as a key negative regulator of iTreg differentiation, providing a novel target and mechanistic insight for optimizing iTreg-based cellular therapies.

1. Research Background: The “Stability Dilemma” of iTreg Cells
Naturally occurring regulatory T cells (nTregs) develop and mature in the thymus, exhibiting stable FOXP3 expression. In contrast, in vitro–induced regulatory T cells (iTregs)—generated from naïve CD4⁺ T cells via TGFβ and IL-2 induction—frequently lose function due to fluctuations in FOXP3 expression. This instability stems from the complexity of the FOXP3 regulatory network, in which the TGFβ-SMAD signaling pathway and epigenetic modifications (e.g., CNS2 region DNA methylation) are known to play roles. However, the key regulators across the genome remain undefined.
The authors note that previous research has primarily focused on the maintenance mechanisms of nTregs, whereas the molecular mechanisms governing iTreg differentiation have yet to be systematically elucidated. Therefore, employing unbiased genome-wide screening to identify critical genes influencing FOXP3 induction represents a pivotal strategy for overcoming the stability barrier in iTreg-based therapies.
2. Technical Core: A “High-Precision Search” via Genome-Wide CRISPR Screening
To systematically identify FOXP3 regulatory genes, the research team developed a CRISPR screening platform based on SLICE( sgRNA lentiviral infection with Cas9 electroporation), effectively functioning as a “genomic-level microscope.”
(1) Screening design: Targeting cell populations with differential FOXP3 expression
Using a lentiviral sgRNA library targeting 19,113 genes (77,491 sgRNAs, four per gene), human primary naïve CD4⁺ T cells were transduced and subsequently induced to differentiate into iTregs with TGFβ and IL-2.After 72 hours, cells were separated into FOXP3^high and FOXP3^low populations by flow cytometry. Enrichment differences in sgRNAs between the two populations were analyzed using the MAGeCK algorithm to pinpoint FOXP3 regulators.
The screening not only recovered known regulators such as TGFBR1 and SMAD3, confirming the robustness of the approach, but also identified novel candidates including SMARCB1 and MIDN. Notably, RBPJ emerged as a prominent negative regulator—its sgRNAs were significantly enriched in FOXP3^high cells, suggesting that RBPJ loss enhances FOXP3 expression (Fig. 1b,c).
(2) Validation strategy: From screening to single-gene functional confirmation
To exclude false positives, the team performed single-gene knockout validation using Cas9-gRNA ribonucleoprotein (RNP) delivery. RBPJ deletion led to a marked increase in both the proportion of FOXP3⁺ cells and their mean fluorescence intensity (MFI), with consistent trends observed across five independent donors, demonstrating the robustness of its regulatory role (Fig. 1e,g).
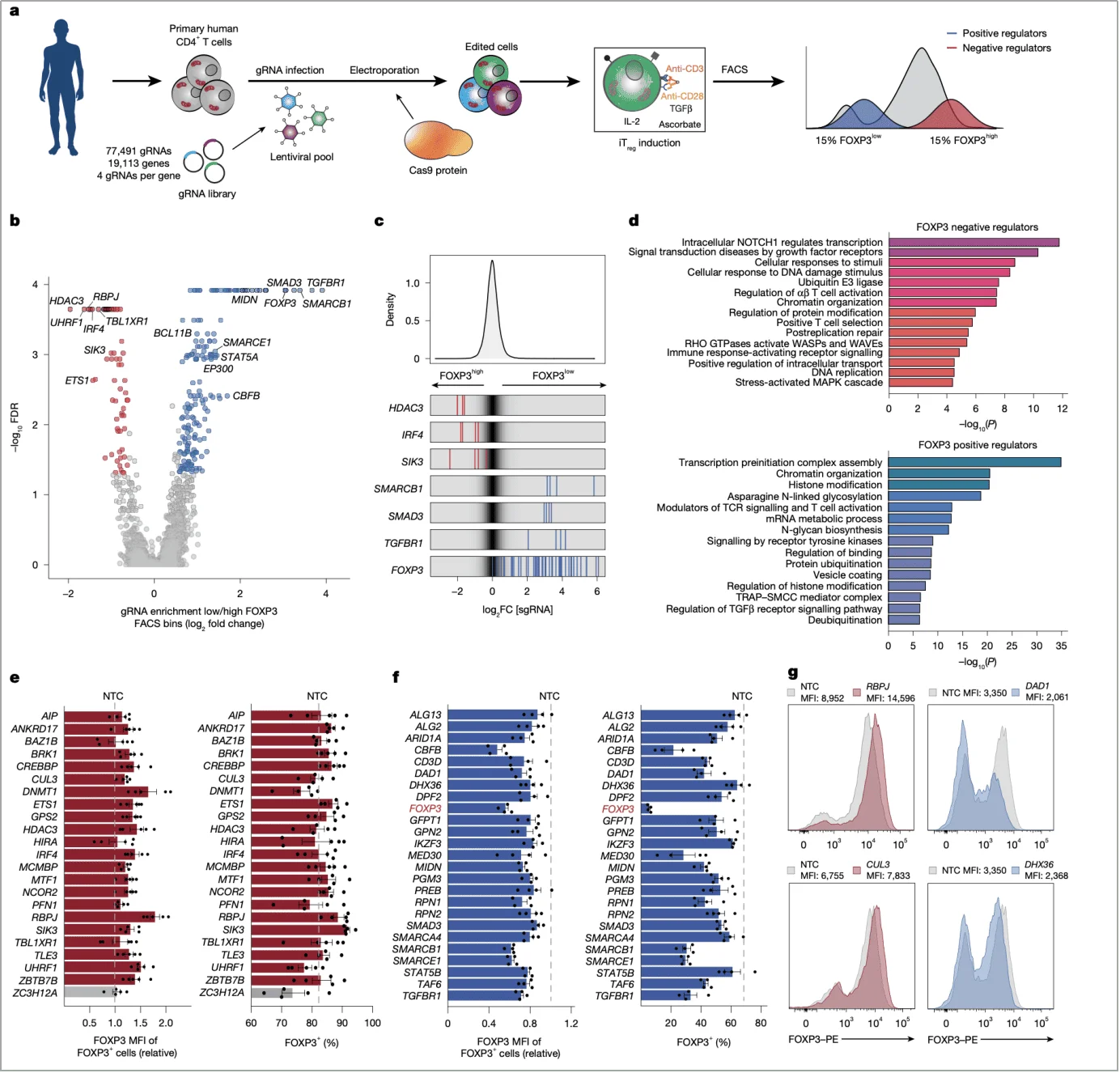
Fig. 1 | A whole-genome CRISPR screen in primary human T cells uncovers novel regulators of FOXP3 induction
3. Mechanistic insights: How does RBPJ “repress” FOXP3 expression?
As a canonical downstream effector of the Notch pathway, RBPJ was unexpectedly found to regulate FOXP3 in a manner independent of Notch signaling—an observation that contradicts the prevailing paradigm.
(1) “Acting independently” of Notch
Perturbation of key Notch pathway components, such as NOTCH1 and MAML1, had no significant effect on FOXP3 expression. In contrast, RBPJ knockout selectively enhanced FOXP3 expression in iTregs, but not in nTregs, indicating a cell type–specific regulatory mechanism that is uncoupled from Notch signaling (Fig. 3b).
(2) “Cooperative repression” with the NCOR–HDAC3 complex
Further investigation revealed that RBPJ, via its β-trefoil DNA-binding domain, associates with the NCOR (nuclear receptor corepressor)–HDAC3 complex to directly target the FOXP3 promoter. ChIP-seq analysis confirmed that RBPJ deletion increased levels of H3ac and H3K9ac-histone modifications associated with transcriptional activation—at the FOXP3 locus. Moreover, pharmacological inhibition of HDAC3 reversed the FOXP3 suppression mediated by RBPJ overexpression, supporting a model in which RBPJ represses FOXP3 transcription through histone deacetylation (Fig. 2f,g).
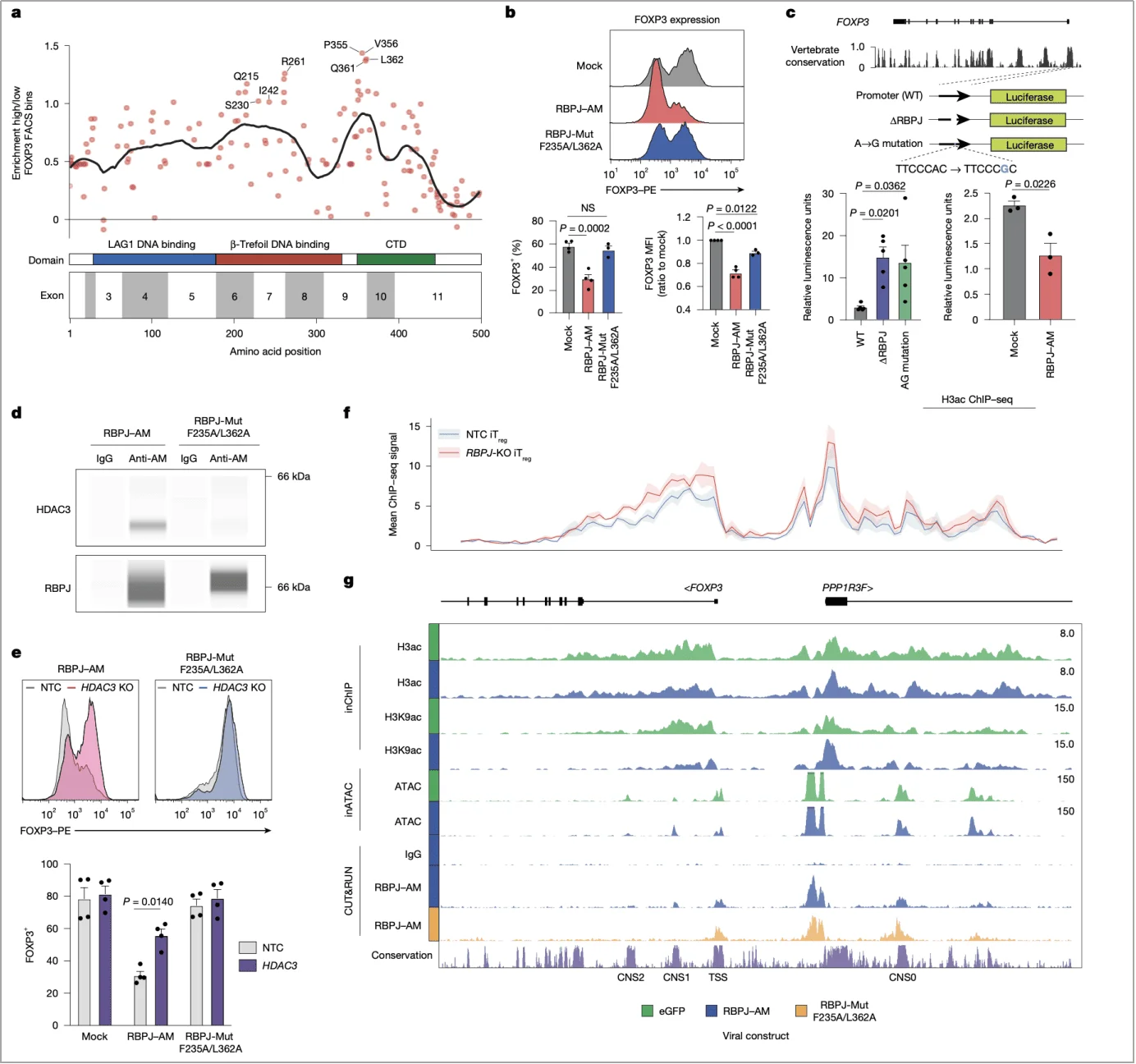
Fig. 2 | The RBPJ–NCOR–HDAC3 complex directly represses FOXP3 through modulation of local histone acetylation.
(3) Epigenetic “dual regulation”
In addition to directly repressing transcription, RBPJ also impacts the epigenetic stability of FOXP3. RBPJ deletion markedly reduced CpG methylation within the CNS2 region of the FOXP3 locus—a region associated with long-term FOXP3 expression. This demethylation was vitamin C–dependent, suggesting that RBPJ may maintain CNS2 methylation by suppressing the activity of DNA demethylases (Fig. 3g).
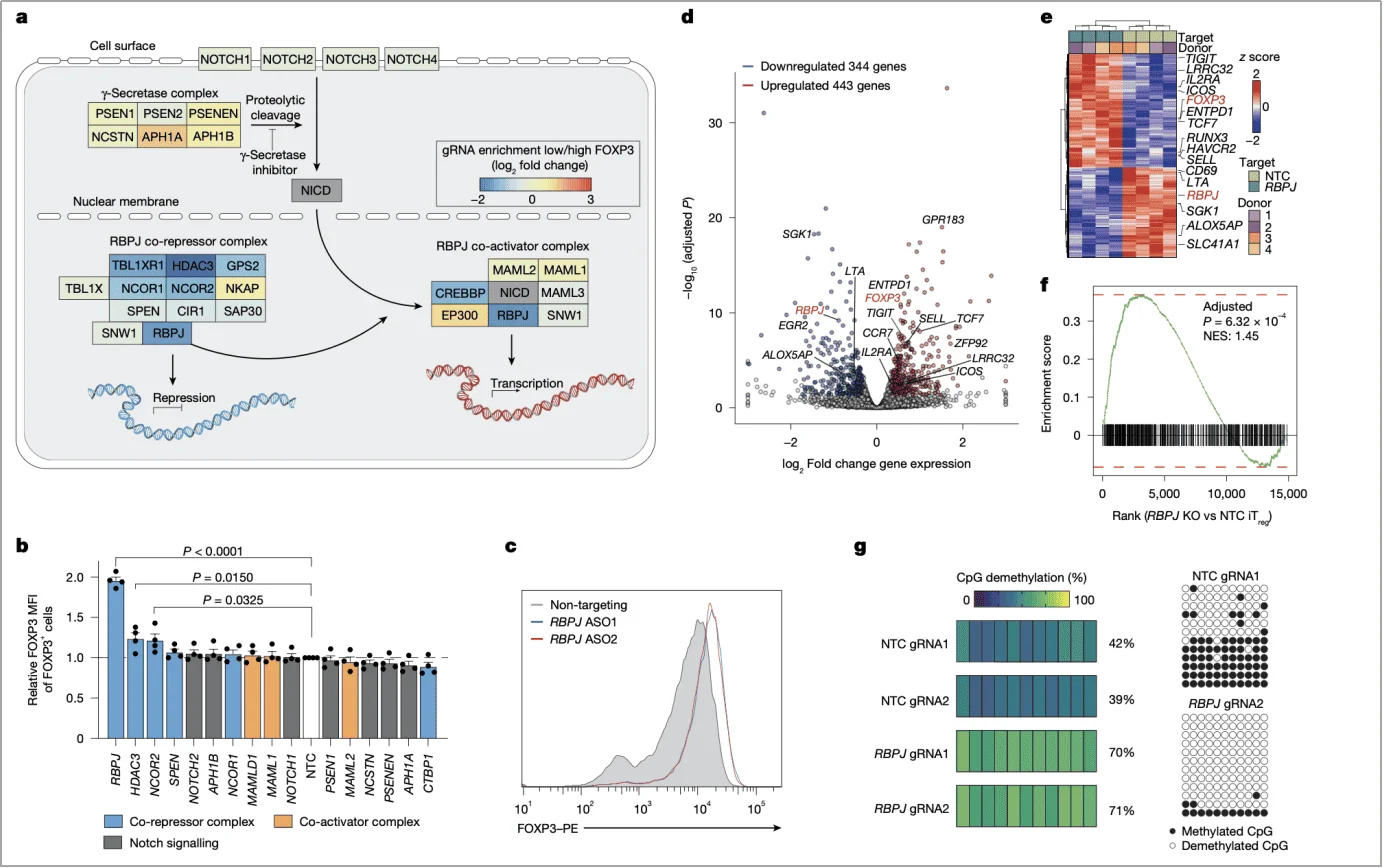
Fig. 3 | RBPJ knockout improves FOXP3 expression, function and stability in iTreg cells.
4. Functional extension: Perturb-icCITE-seq reveals the regulatory network
To delineate the global regulatory effects of RBPJ, the team employed Perturb-icCITE-seq—a platform that integrates CRISPR perturbations with single-cell protein and transcriptome profiling enabling simultaneous profiling of transcriptomic changes following knockout of 296 candidate genes and alterations in the expression of over 300 proteins.
Upon RBPJ deletion, iTregs exhibited upregulation of core functional genes such as IL2RA and ENTPD1, along with increased protein levels of immunosuppressive molecules (CTLA4, CD39), suggesting enhanced suppressive function. Co-functional module analysis classified RBPJ into module H, which displayed distinct regulatory features from the NCOR complex (module I), further supporting its independent mode of action (Fig. 4d,e).
Importantly, this approach also revealed a strong correlation between FOXP3 protein abundance and sgRNA enrichment (r = –0.71), indicating that RBPJ-mediated regulation of FOXP3 is both direct and specific (Fig. 4c).
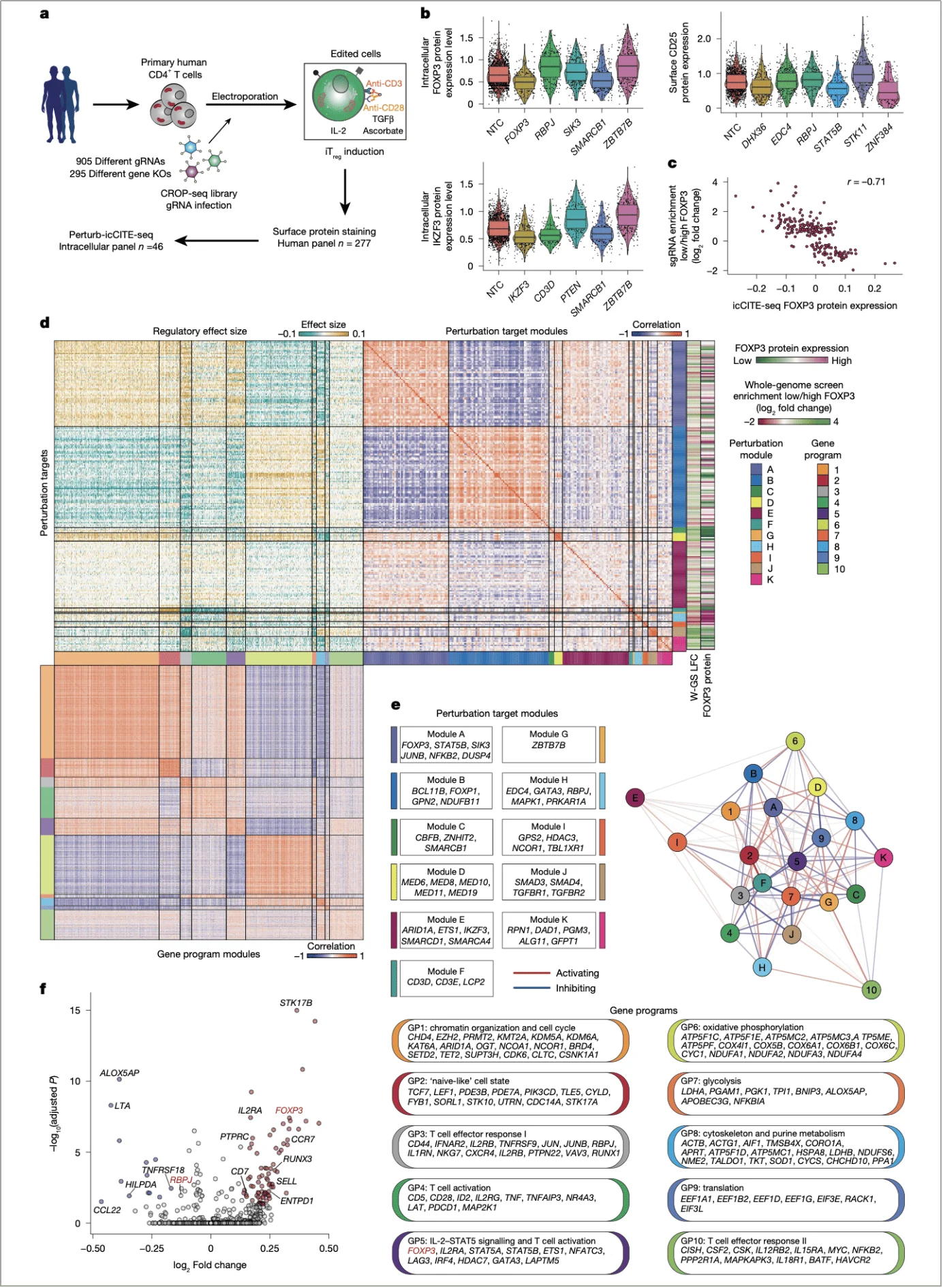
Fig. 4 | Validation of FOXP3 regulators with Perturb-icCITE-seq.
5. Clinical potential: Functional “boost” of RBPJ-deficient iTregs
From mechanistic dissection to preclinical validation, the study demonstrated that RBPJ deletion markedly enhances the therapeutic potential of iTregs.
(1) In vitro function: Dual improvement in stability and suppressive capacity
Under repeated TCR stimulation and TNF (inflammatory cytokine) challenge, the proportion of FOXP3⁺ cells in RBPJ-deficient iTregs remained above 70%, significantly higher than the ~40% observed in controls. Suppression assays revealed a 2–3-fold increase in their ability to inhibit the proliferation of both CD4⁺ and CD8⁺ effector T cells, with suppression still effective at a low iTreg-to-responder ratio of 1:16 (Fig. 5).
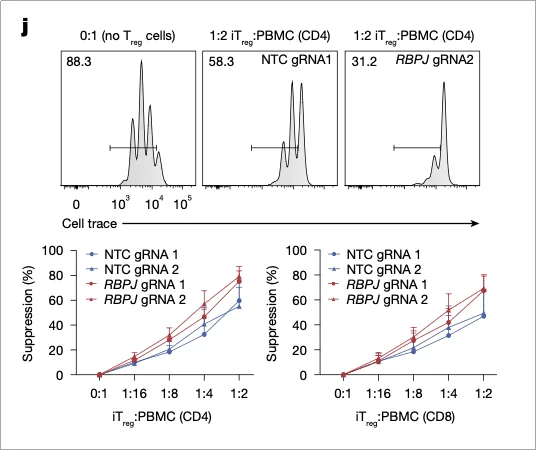
Fig. 5 | Suppression assay confirms enhanced suppressive activity of RBPJ-deficient iTregs.
(2) In vivo validation: Demonstrating therapeutic advantage in a GvHD model
In a humanized mouse model of graft-versus-host disease (GvHD), mice receiving RBPJ-deficient iTregs exhibited an 80% survival rate, significantly higher than controls—approximately 70% survival with nTreg infusion and only 30% with control iTregs. Additionally, these mice experienced less weight loss, and after 5 days, 91.5% of infused cells maintained high FOXP3 expression, demonstrating their sustained in vivo stability (Fig. 6b,c,d).
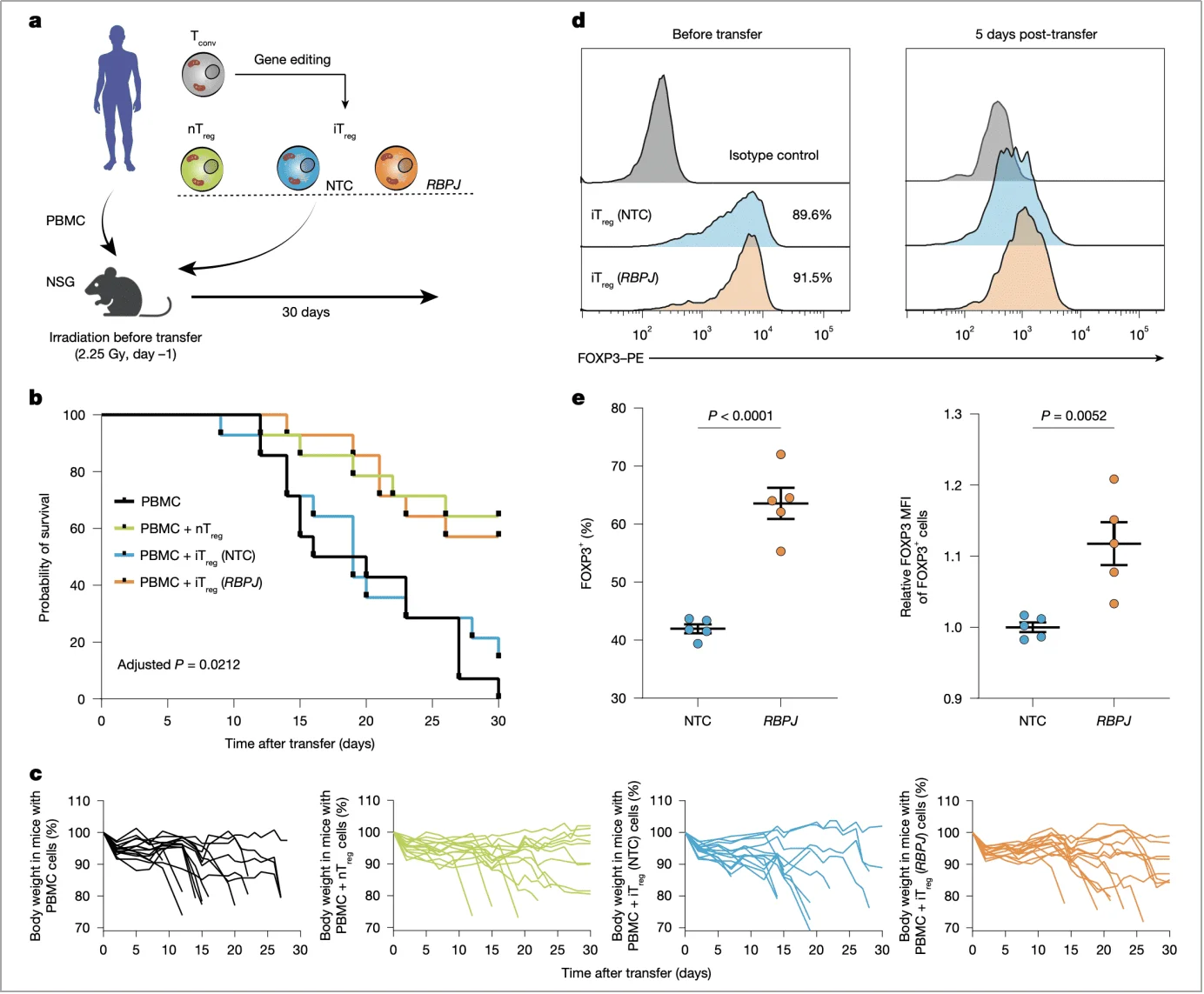
Fig. 6 | RBPJ ablation improves iTreg in vivo stability and suppressive function.
6. Technological innovations: Precision tools for FOXP3⁺ cell–focused analyses
To overcome the limitations of conventional methods that struggle to specifically target distinct cell populations, the research team developed three novel technologies, providing a targeted perspective for epigenetic and transcriptomic profiling:
- · icRNA-seq: RNA extraction following flow cytometric sorting of FOXP3⁺ cells enables precise transcriptomic analysis, minimizing interference from FOXP3⁻ cells;
- · inChIP-seq: Chromatin immunoprecipitation performed specifically on FOXP3⁺ cells to uncover the impact of RBPJ on histone modifications at the FOXP3 promoter;
- · inATAC-seq: Assessment of chromatin accessibility in FOXP3⁺ cells reveals that RBPJ deficiency enhances accessibility at both the FOXP3 promoter and CNS2 regions.
These approaches overcome the challenge of mixed-cell population analyses that obscure critical signals establishing a paradigm for investigating cell type–specific regulation of other transcription factors.
7. Significance and outlook: Bridging basic research and cell therapy
This study not only uncovers a novel regulatory mechanism of FOXP3, but also provides practical strategies to optimize iTreg cell therapy:
- · Theoretical breakthrough: Identification of RBPJ as a novel regulator enriches the epigenetic regulatory network of FOXP3 and explains the stability differences between iTregs and nTregs.
- · Clinical potential: RBPJ deletion enhances iTreg stability and function, offering a promising new avenue for cell therapies targeting autoimmune diseases such as rheumatoid arthritis and organ transplant rejection.
- · Technological inspiration: The integration of genome-wide CRISPR screening with single-cell multi-omics establishes a versatile paradigm for investigating cell fate regulation in other contexts.
From a genome-wide “carpet bombing” approach to a comprehensive upgrade in cellular function, this work brings us closer to the goal of controlled, stable iTreg therapies. In the near future, iTregs optimized via RBPJ modulation may serve as precisely trained “peacekeepers,” vigilantly safeguarding our immune system and offering new hope to countless patients.
At Ubigene, we've built our own CRISPR-iScreen™ platform to support a wide range of research needs. With a complete system for phenotypic screening and data analysis, we help you every step of the way—from planning your experiments to digging into the results analyzing the results —so your research moves forward smoothly and faster. Whether you need CRISPR library plasmids、CRISPR library viruses、CRISPR library cell pools, or help setting up screening workflows and NGS sequencing, we provide comprehensive one-stop, end-to-end CRISPR screening services. Our goal is to make sure every part of your project fits together perfectly, making your work easier and more efficient.
Ubigene is about to launch a brand-new automated CRISPR library analysis platform that fully automates bioinformatics data processing. Researchers won't need to rely on outside services external services anymore—they can easily handle CRISPR screening data analysis and result visualization right in their own labs. This will greatly boost work efficiency, save personnel resources and costs, and truly put the data processing “in your own hands,” making research more efficient and flexible than ever before.
For more information or technical support, please don't hesitate to contact us>>
Reference
Chen, K.Y., Kibayashi, T., Giguelay, A. et al. Genome-wide CRISPR screen in human T cells reveals regulators of FOXP3. Nature 642, 191–200 (2025).


