CRISPR Functional Genomics Reveals Senolytic Dependencies in Senescent Cells


In life science research, cellular senescence is increasingly being redefined—from a passive “end point” of proliferation to an actively regulated cellular state with profound biological functions. A growing body of evidence indicates that during physiological aging, as well as in contexts such as cancer therapy, chronic inflammation, and tissue injury, senescent cells are not efficiently eliminated. Instead, they can persist long term while continuously secreting a complex mixture of pro-inflammatory cytokines, growth factors, and extracellular matrix-remodeling proteins, collectively referred to as the senescence-associated secretory phenotype (SASP).
While SASP factors may exert transient tumor-suppressive or tissue-protective effects, the chronic accumulation of senescent cells ultimately drives tissue dysfunction, promotes tumor relapse, and fuels therapeutic resistance. Consequently, the selective elimination of senescent cells—through so-called senolytic strategies—has emerged as a highly promising approach in both aging research and oncology.
However, the central challenge is not simply whether senescent cells can be forced into cell death, but rather a more fundamental question: why are senescent cells able to survive long term in an environment that is otherwise strongly pro-apoptotic? To systematically dissect the survival mechanisms that underpin the senescent state, the laboratory of René Bernards provided a paradigmatic example in a study published in Nature Cancer in 2022. The significance of this work lies not primarily in the specific targets identified, but in its conceptual framework: a CRISPR-based functional genomic screening strategy explicitly designed to uncover senescence-state-specific dependencies. This approach reframes senolytic target discovery around the unique vulnerabilities that sustain senescent cell survival, offering a powerful roadmap for the rational development of next-generation senolytic therapies.

Figure 1. Literature search for articles on aging research
Screening Logic: Identifying the targets Among 20,000 Candidates
The study was built around a central question: under continuous exposure to SASP-associated stress signals, which molecular mechanisms do senescent cells rely on to avoid activating apoptotic programs?
To address this, the researchers employed a genome-wide CRISPR knockout library and performed negative selection screens in human cancer cell models (A549 and SK-HEP-1). Cells were first driven into a stable senescent state using pharmacological agents, and the CRISPR screen was then conducted specifically within this defined cellular context.
To minimize bias introduced by any single senescence-induction mechanism, multiple mechanistically distinct triggers were applied in parallel, including the Aurora kinase A inhibitor alisertib, the PLK4 inhibitor CFI-400945, and the topoisomerase II inhibitor etoposide. Only genes that consistently showed depletion across different cell models and across all senescence-inducing conditions were classified as bona fide senescence dependency factors. In other words, the screen was not designed to identify “senescence-associated genes,” but rather to uncover survival factors that senescent cells cannot afford to lose.
The conceptual strength of this design lies in its focus on non-redundant survival dependencies that are uniquely required in the senescent state. By anchoring the screen to functional indispensability rather than differential expression, the approach effectively minimizes confounding effects arising from acute stress responses, transient DNA damage, or stochastic transcriptional fluctuations.
Within this framework, the screen identified an unexpected yet robust hit: cFLIP (CFLAR) — a gene not traditionally central to senolytic research, but one that exhibited a pronounced survival dependency specifically in senescent cells. cFLIP is a well-established inhibitor of the extrinsic apoptotic pathway, acting by preventing formation of the FADD-caspase-8 death-inducing signaling complex (DISC) and thereby suppressing death receptor-mediated apoptosis. Importantly, its relevance in this study does not stem from its known molecular function per se, but from the heightened and non-redundant dependency that emerges uniquely within the physiological context of cellular senescence.
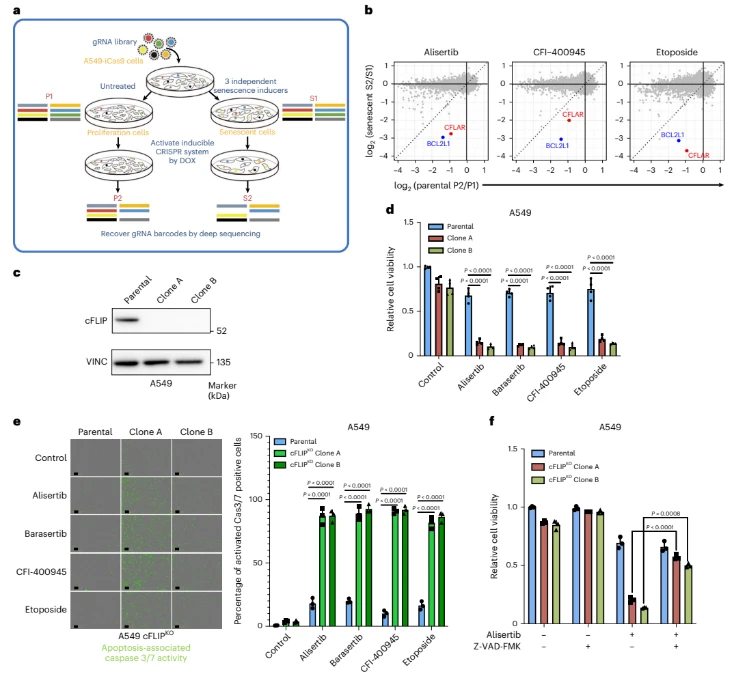
Figure 2. Survival-Based CRISPR Screening Strategy
"Senescent Cells at Full Speed": The Tug-of-War Between Accelerator and Brake
Subsequent mechanistic analyses revealed that senescent cells exist in a state of extreme tension—an unstable equilibrium that can be captured by a highly simplified yet powerful conceptual model. On one hand, persistent SASP-driven signaling leads to chronic activation of the NF-κB pathway, resulting in marked upregulation of the death receptor DR5 and its ligand TRAIL. As a consequence, the extrinsic apoptotic machinery is maintained in a primed, hypersensitive state. Metaphorically, senescent cells have the apoptotic “accelerator” fully depressed, poised to self-destruct at any moment. On the other hand, to counterbalance this continuous pro-apoptotic pressure, senescent cells simultaneously and robustly upregulate cFLIP expression. Acting as a critical negative regulatory node, cFLIP blocks signal transmission from DR5 to caspase-8, thereby preventing execution of death receptor-mediated apoptosis.
This condition can be summarized as an extreme “accelerator-and-brake” equilibrium: apoptotic signals accumulate continuously but are narrowly restrained by cFLIP. Once this inhibition is relieved, cells rapidly cross the apoptotic threshold and undergo cell death.
Secondary Screening: Bridging Toward Clinical Translation
Despite these insights, the Bernards team recognized that primary hits from functional genomics screens remain several steps removed from clinical application. Directly targeting cFLIP alone poses challenges related to potential toxicity and limited therapeutic selectivity.
To address this, the investigators implemented a secondary screening strategy — not to identify an additional dependency per se, but to determine which regulatory nodes might be more amenable to safe and controllable pharmacological intervention. They assembled a focused CRISPR library enriched for kinases and epigenetic regulators and screened for genes that exhibited synthetic lethality when combined with DR5 agonists.
This approach led to the identification of BRD2. Inhibition of BRD2 not only suppresses cFLIP transcription, but also remodels chromatin accessibility to enhance the transcriptional responsiveness of pro-apoptotic genes. Together, these effects release the previously restrained apoptotic program and convert a fragile equilibrium into an irreversible death signal.Conceptually, this transforms a single, vulnerable strike into a coordinated combination therapy: cytotoxic chemotherapy is first used to drive tumor cells into a senescent state, followed by targeted elimination through the combined use of BRD2 inhibitors and DR5 agonists. In patient-derived xenograft (PDX) models, this sequential therapeutic strategy produced striking tumor regression, highlighting its translational potential.
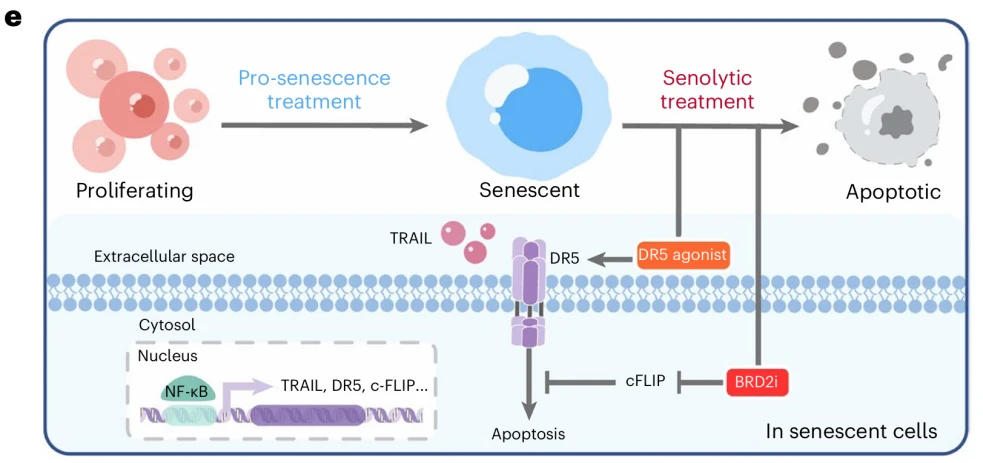
Figure 3
If we step back from individual targets, the most enduring value of this study lies in a clear, transferable screening logic rather than in any single molecular finding.
A Practical Playbook for Researchers: Reusable Methodology
In retrospect, the impact of this work extends well beyond the identification of the cFLIP–DR5 senolytic axis. Its true contribution is the demonstration of a highly generalizable research paradigm.
First, the foundation of functional genomics screening is a precise definition of cellular state, not a simple comparison of gene knockout effects. The most informative hits emerge from contrasts in which cells survive under control conditions but become nonviable only within a specific physiological state. It is this state-dependent lethality that reveals genuine biological dependencies.
Second, negative-selection (dropout) screens have a unique advantage in uncovering survival dependencies. Compared with positive-enrichment strategies, depletion-based screens more directly expose non-redundant regulatory nodes that are indispensable under defined conditions.
Third, the purpose of secondary screening is not merely to refine mechanistic understanding, but to identify intervention strategies that are combinable and pharmacologically tractable. As research moves toward clinical translation, synergistic targets within epigenetic or signaling networks often offer greater therapeutic feasibility than direct inhibition of a single core survival factor.
Conclusion: Finding the True Switch Within Complexity
Biological systems are inherently redundant, adaptive, and dynamically rewired—senescent cells perhaps most of all. The true power of functional genomics screening does not lie in exhaustively mapping every pathway, but in systematically perturbing the system to pinpoint the critical nodes that sustain survival.
The discovery of the cFLIP-DR5 axis underscores a broader principle: the strongest forms of resistance often indicate that a system has been compressed onto a narrow, irreversible survival pathway.
By placing cellular state at the center of screening logic, functional genomics is opening more precise and controllable entry points for intervening in cells that are, quite literally, old but unwilling to die.
Ubigene CRISPR Library Screening Services
Ubigene provides end-to-end CRISPR library screening services for both in vitro and in vivo applications. We design customized screening strategies tailored to specific scientific questions. Whether your research focuses on immune-tumor interactions, mechanisms of drug resistance, cancer metastasis, or systemic phenotypic effects, Ubigene delivers integrated solutions spanning experimental design, execution, and comprehensive data interpretation.Supported by robust phenotypic screening workflows and an advanced data analytics infrastructure, Ubigene enables high-resolution functional interrogation to accelerate both basic discovery and translational research.
Ubigene has also independently developed an interactive data analysis solution — the iScreenAnlys™ CRISPR Library Analysis Platform. Designed with an intuitive, user-friendly interface, iScreenAnlys™ allows researchers to perform end-to-end CRISPR library data analysis with no prior bioinformatics expertise required. The platform integrates three widely adopted algorithms — DrugZ, MAGeCK-RRA, MAGeCK-MLE and supports multiple statistical methods with flexible, publication-ready visualization outputs.
Contact us to learn more >>>Reference
[1] Wang, L., Jin, H., Jochems, F. et al. cFLIP suppression and DR5
activation sensitize senescent cancer cells to senolysis. Nat Cancer 3,
1284–1299 (2022).
[2] Childs, B., Gluscevic, M., Baker, D. et al. Senescent cells: an
emerging target for diseases of ageing. Nat Rev Drug Discov 16, 718–735
(2017).
[3] Wu Z, Qu J, Zhang W; Aging Biomarker Consortium; Liu GH. Biomarkers
of ageing of humans and non-human primates. Nat Rev Mol Cell Biol. 2025
Nov;26(11):826-847.


