IF 19.8 | GPNMB Disrupts SNARE Complex Assembly to Promote Intracellular Bacterial Survival


Introduction
Xenophagy plays a critical role in restricting bacterial proliferation within macrophages. However, the molecular mechanisms governing autophagosome–lysosome fusion during bacterial infection remain incompletely understood.
On March 4, 2025, Zhenzhen Yan et al. published a research article in Cellular & Molecular Immunology (CMI, Impact Factor: 19.8) entitled “GPNMB disrupts SNARE complex assembly to maintain bacterial proliferation within macrophages.” This study uncovers a previously unrecognized mechanism by which GPNMB promotes the survival and replication of intracellular bacteria in macrophages by disrupting the assembly of the SNARE complex during infection. Notably, the GPNMB knockout THP-1 cell line used in this study was generated and supplied by Ubigene, providing a critical experimental tool that enabled the mechanistic dissection of GPNMB function in host-pathogen interactions.
Research Background
Leprosy is a chronic infectious disease caused by Mycobacterium leprae (M. leprae). Macrophages are the primary host cells in which M. leprae resides and replicates, and functional impairment of these cells is a key factor enabling intracellular bacterial survival and proliferation. However, the molecular mechanisms that regulate immune evasion by M. leprae remain poorly understood.
During microbial infection, autophagy serves as a critical component of the host immune defense, contributing to the elimination of intracellular pathogens. By adapting to or subverting host autophagic pathways, pathogens can establish persistent infections and, in some cases, induce host cell death. Previous studies have shown that inactivated M. leprae can induce autophagy in human monocytes, whereas live M. leprae actively suppresses this process. These findings indicate that inhibition of autophagy is a central strategy employed by M. leprae to evade host immune responses, although the precise molecular mechanisms underlying this suppression have yet to be elucidated.
Research Objectives
- Clarify the role of GPNMB in intracellular bacterial infection and elucidate its association with immune suppression and bacterial proliferation in patients with lepromatous leprosy (L-Lep).
- Elucidate the molecular mechanisms by which GPNMB regulates host antibacterial immunity, with a focus on its impact on autophagy pathways and SNARE complex assembly.
- Assess the feasibility of targeting GPNMB as a potential therapeutic strategy for diseases associated with intracellular bacterial infections.
Research Methods
Sample Analysis: GPNMB expression was assessed in skin tissues from leprosy patients (L-Lep and T-Lep) using multiplex immunohistochemistry (mIHC), and serum levels of soluble GPNMB were measured by ELISA. Expression differences were further validated using publicly available databases.
Cellular Experiments: Wild-type and GPNMB knockout THP-1 cells, as well as HEK293T cells, were used to manipulate gene expression via plasmid overexpression or siRNA knockdown. Protein-protein interactions were examined by co-immunoprecipitation (Co-IP), while expression levels of key proteins (LC3, p62, SNAP29, etc.) were measured by Western blot. Autophagosome-lysosome colocalization was visualized through immunofluorescence microscopy. Multiple intracellular bacterial infection models were established to evaluate cytokine secretion (qPCR/ELISA) and bacterial burden.
Animal Experiments: Conditional Gpnmb knockout mice (Gpnmb^fl/fl Lyz2-Cre) and wild-type controls were infected with Mycobacterium marinum to assess in vivo bacterial burden, tissue pathology, and cytokine levels.
Molecular Mechanism Validation: STX17 truncation mutants and GPNMB glycosylation site mutants were constructed. Functional effects on autophagy and SNARE complex assembly were evaluated using a GFP-mCherry-LC3 dual-fluorescence reporter system, allowing precise mapping of protein interaction domains and post-translational modifications.
Research Workflow
Clinical Correlation Analysis: High GPNMB expression in L-Lep patients was correlated with weakened immune responses and elevated bacterial loads, identifying a key scientific question regarding its role in intracellular bacterial survival.
Cellular Functional Validation: Using GPNMB knockout and overexpression cell models, the effects on host immune responses, bacterial proliferation, and autophagic flux were assessed. The functional impact was localized specifically to the autophagosome-lysosome fusion step.
Molecular Mechanism Investigation: GPNMB was found to specifically interact with STX17. Mechanistically, GPNMB induces its own deglycosylation and promotes SNAP29 degradation, thereby blocking SNARE complex assembly and preventing autophagosome–lysosome fusion.
In Vivo Validation: Mouse infection models confirmed that GPNMB knockout enhances bacterial clearance and strengthens immune responses, validating the cellular findings in an organismal context.
Integrated Conclusion: A complete mechanistic pathway was established illustrating how GPNMB regulates intracellular bacterial infection, highlighting its potential as a therapeutic target.
Research Highlights
1. Elevated GPNMB Expression in Lepromatous Leprosy (L-Lep)
In this study, the authors compared GPNMB levels in skin tissues and blood samples between lepromatous leprosy (L-Lep) patients—characterized by weak immune responses and high bacterial loads—and tuberculoid leprosy (T-Lep) patients, who exhibit strong immunity and low bacterial burden. They found that GPNMB levels were significantly higher in L-Lep patients, suggesting a potential role of GPNMB in facilitating M. leprae immune evasion.
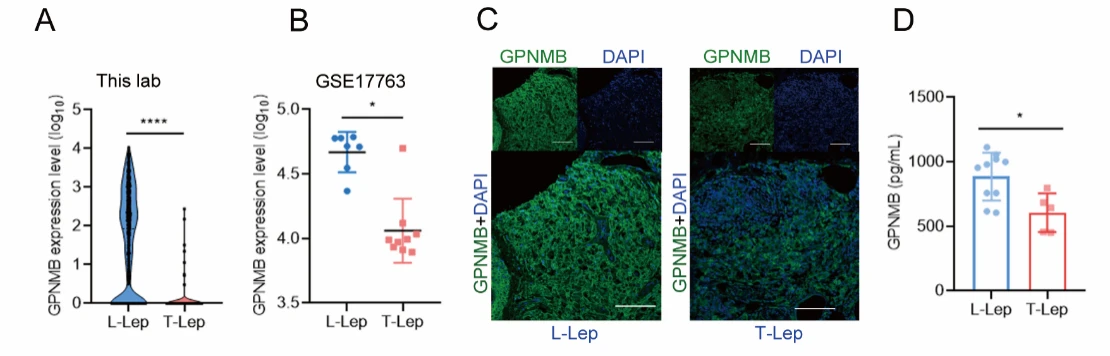
Figure 1. GPNMB Levels Are Significantly Higher in Lepromatous Leprosy (L-Lep) Patients Compared with Tuberculoid Leprosy (T-Lep)
2. GPNMB Promotes Intracellular Bacterial Survival
To investigate the functional role of GPNMB, the authors knocked down GPNMB expression in macrophages using siRNA. Cells with reduced GPNMB levels exhibited significantly higher intracellular bacterial loads. Similarly, in a Mycobacterium marinum infection mouse model, GPNMB knockout mice showed markedly increased bacterial burdens across multiple organs. These findings demonstrate that GPNMB facilitates the survival of intracellular bacteria.
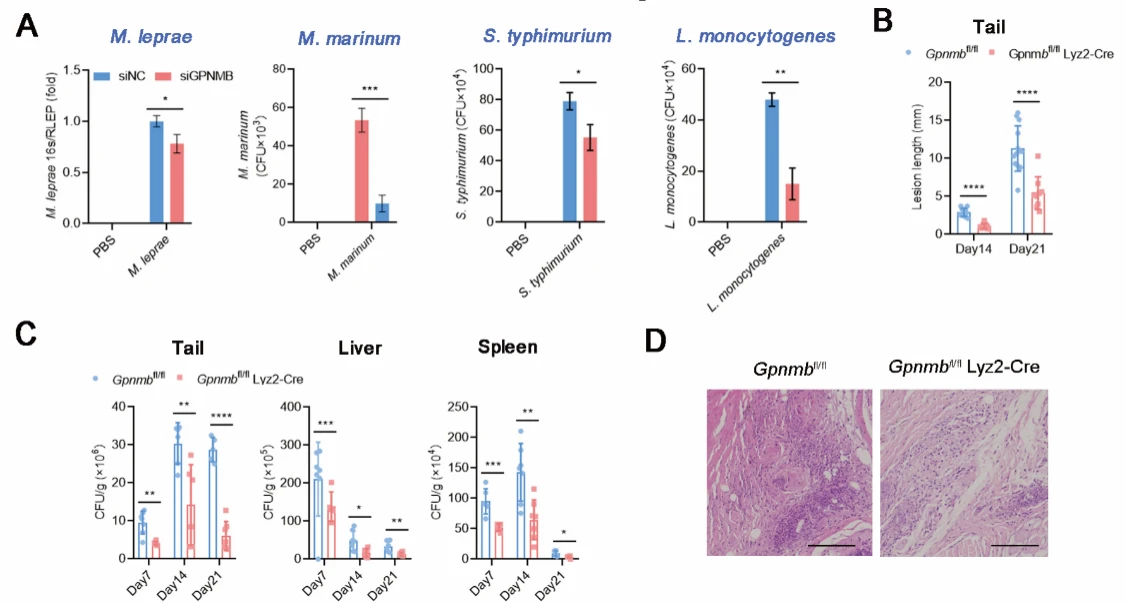
Figure 2. GPNMB Facilitates the Survival of Intracellular Bacteria
3. GPNMB Inhibits Cellular Autophagy
To elucidate the mechanism of GPNMB, the authors examined its subcellular localization and found that during bacterial infection, GPNMB clearly colocalizes with autophagosome markers LC3 and p62. In GPNMB knockout cells, the levels of LC3 and p62 were significantly reduced, indicating that GPNMB disrupts the autophagic process.
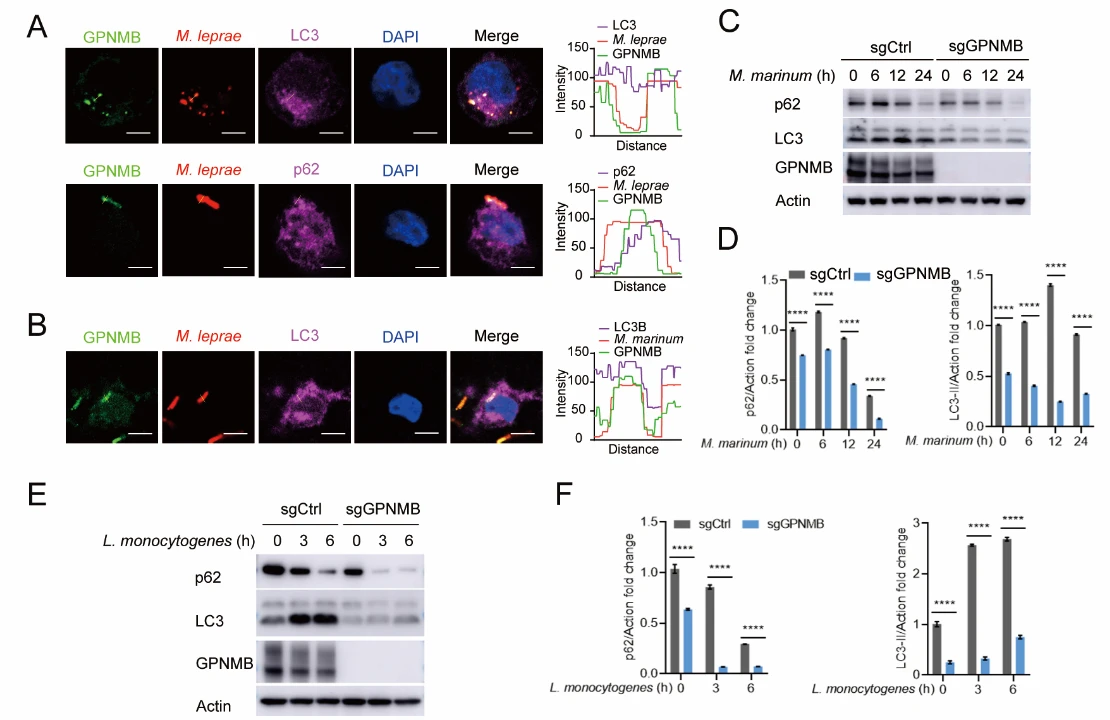
Figure 3. GPNMB Inhibits Cellular Autophagy
4. GPNMB Inhibits Autophagosome-Lysosome Fusion
To investigate the mechanism by which GPNMB regulates autophagy, the authors conducted a series of experiments and found that GPNMB suppresses the fusion of autophagosomes with lysosomes, thereby blocking autophagic flux.
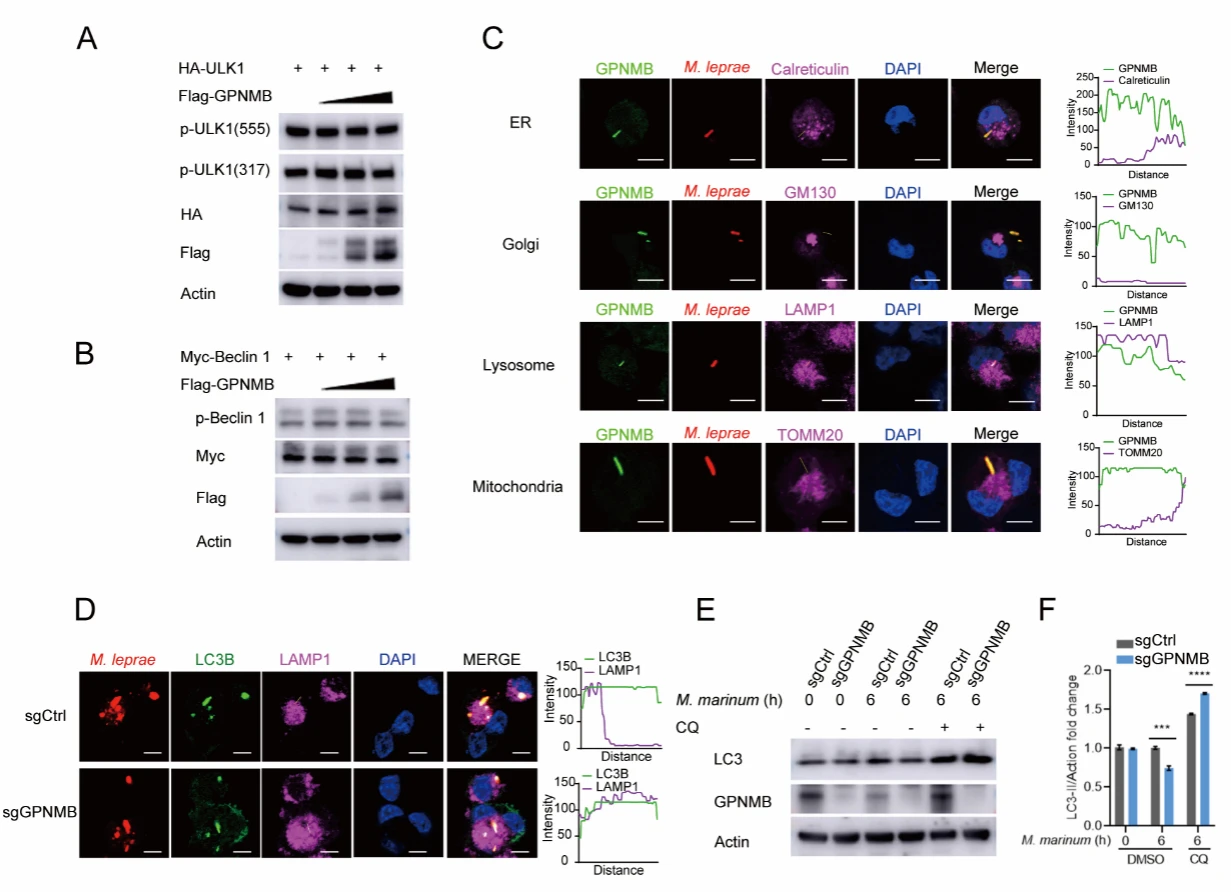
Figure 4. GPNMB Inhibits Autophagosome–Lysosome Fusion
5. GPNMB Specifically Interacts with STX17
Subsequently, the authors screened for molecular targets of GPNMB and identified a specific interaction with the lysosome-associated protein STX17. Moreover, the membrane-targeting domain of STX17 was found to be critical for this interaction, indicating that proper membrane localization is essential for GPNMB–STX17 binding.
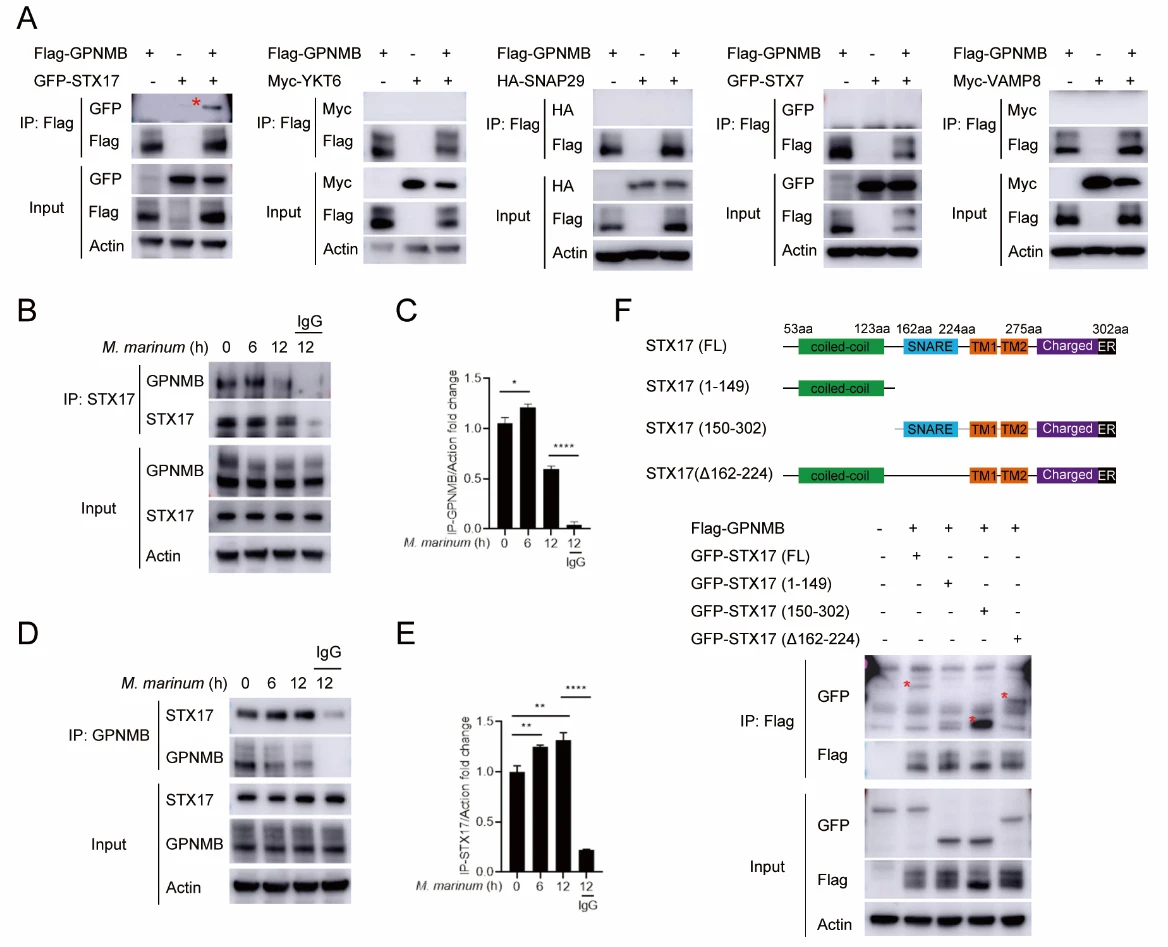
Figure 5. GPNMB Specifically Interacts with STX17
6. GPNMB Inhibits the Interaction Between STX17 and SNAP29
STX17 is a critical component of the STX17-SNAP29-VAMP8 SNARE complex, which mediates autophagosome-lysosome fusion. The authors found that GPNMB disrupts this process by inhibiting the binding between STX17 and SNAP29, thereby blocking SNARE complex assembly and autophagic flux.
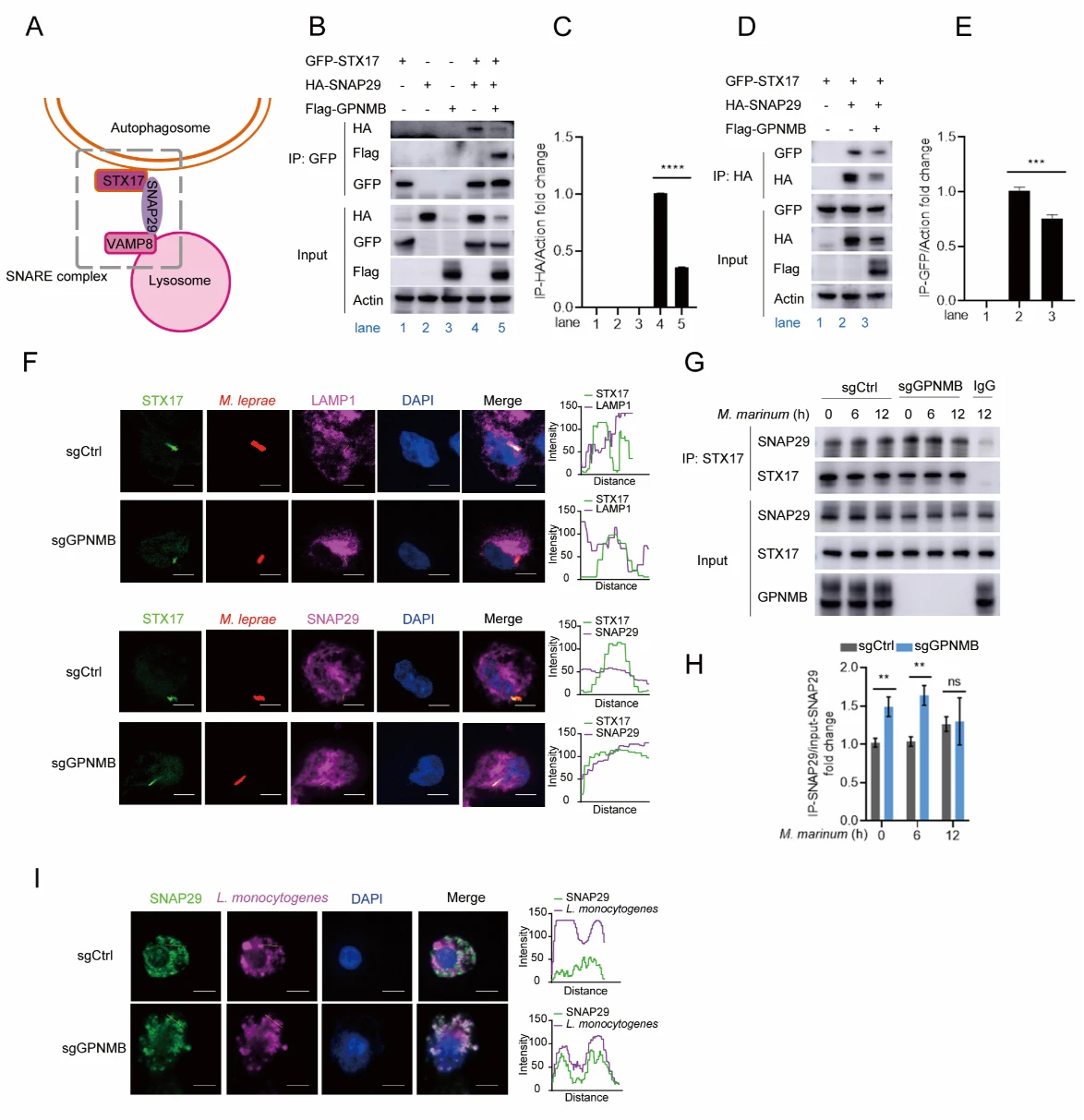
Figure 6. GPNMB Inhibits the Interaction Between STX17 and SNAP29
Significance and Innovations
- Mechanistic Discovery: This study is the first to demonstrate that GPNMB specifically binds STX17 and induces deglycosylation at its N296 site to promote SNAP29 degradation. This disrupts the assembly of the STX17–SNAP29–VAMP8 SNARE complex, inhibits autophagosome–lysosome fusion, weakens host antibacterial immunity, and ultimately facilitates intracellular bacterial proliferation.
- Filling Knowledge Gaps: The work not only elucidates the previously unknown role of GPNMB in intracellular bacterial infection but also clarifies a core mechanism underlying immune suppression in lepromatous leprosy (L-Lep). It establishes a novel axis—GPNMB → autophagy regulation → bacterial survival—providing potential therapeutic targets and biomarkers for chronic intracellular infections.
- Robust Multidimensional Validation: Using a three-tiered validation framework—clinical samples, multiple cell models, and gene knockout mice—the study demonstrates the generalizability of its conclusions across various intracellular bacteria. Importantly, it highlights the critical role of glycosylation modifications in autophagy regulation, offering a new paradigm for investigating host-pathogen interactions and post-translational regulation of autophagy.
Research Conclusions
Leprosy is a classic intracellular bacterial infection and has long served as an ideal model to study the interplay between host defense mechanisms and pathogen survival. Using leprosy as a model, this study systematically reveals a previously unrecognized function of GPNMB in host antibacterial defense. The key findings are as follows: upon bacterial infection, GPNMB is recruited to autophagosomes and interacts with the autophagosome-resident protein STX17 , thereby blocking the assembly of the STX17–SNAP29–VAMP8 SNARE complex. This disruption directly impairs autophagosome–lysosome fusion, inhibits autophagic flux, and ultimately diminishes the host’s ability to clear intracellular bacteria.
Importantly, the effect of GPNMB is not limited to M. leprae. GPNMB deficiency also significantly restricts the intracellular proliferation of multiple pathogens—including M. marinum, Listeria monocytogenes, and Salmonella—in macrophages, indicating a broad regulatory role in intracellular bacterial infections.In vivo studies using a mouse infection model further confirmed these findings: Gpnmb^fl/fl Lyz2-Cre mice exhibited significantly lower bacterial burdens of M. marinum across tissues compared with wild-type controls, underscoring the critical role of GPNMB in facilitating intracellular bacterial survival.
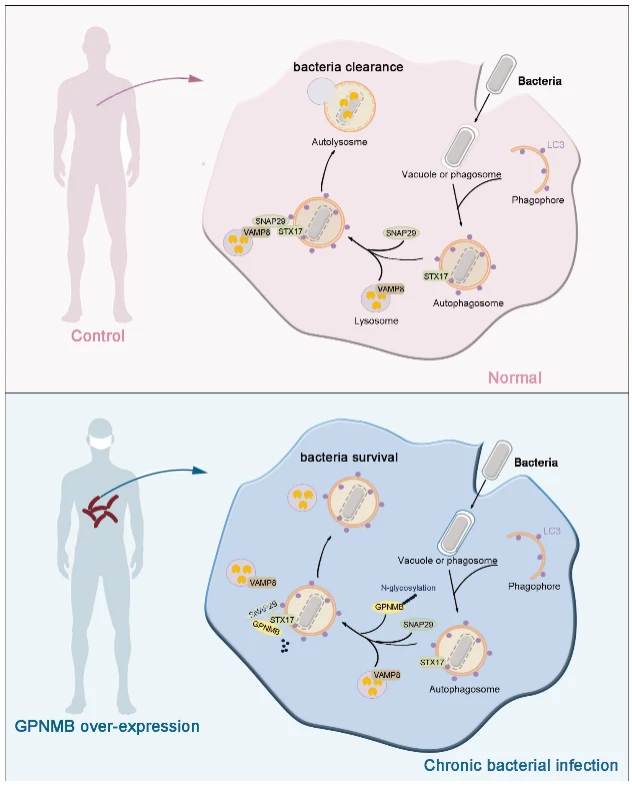
Figure 7. Mechanistic Overview of the Study
This study elucidates, for the first time, that GPNMB regulates host antibacterial defense by modulating SNARE complex assembly, providing a novel conceptual framework for developing autophagy-targeted anti-infective therapies.
Support Provided by Ubigene
In this study, the GPNMB Knockout cell line (THP-1) provided by Ubigene served as a “GPNMB-deficient” control model. By comparing its phenotype with wild-type THP-1 cells, researchers were able to systematically validate the molecular mechanism of GPNMB in regulating intracellular bacterial infection across four key levels: immune response → bacterial survival → autophagic flux → molecular interactions.
Ubigene brings extensive experience in gene editing, with over 13,000 successful cases covering a wide range of popular research targets and classical cell lines. We provide mature gene knockout solutions, including:
- Over 8,000 KO cell lines in stock, starting at ¥4,980. Search for your target in the Red Cotton·Wanxiang Full-Category Cell Bank.
- Customized KO cell construction services tailored to your research needs, supporting efficient exploration in fields such as infection immunology and autophagy regulation.
Reference
Yan Z, Han J, Mi Z, Wang Z, Fu Y, Wang C, Dang N, Liu H, Zhang F. GPNMB disrupts SNARE complex assembly to maintain bacterial proliferation within macrophages. Cell Mol Immunol. 2025 May;22(5):512-526. doi: 10.1038/s41423-025-01272-z. Epub 2025 Mar 4. PMID: 40038549; PMCID: PMC12041529.


