Target Discovery Using In Vivo CRISPR Library Screening


In the era of precision medicine, target identification represents the foundation of drug development, and CRISPR library technology is driving transformative advancements in this domain. Transitioning CRISPR library screening from in vitro to in vivo models not only provides a physiologically relevant environment but also enables the discovery of targets with direct translational potential.
Why In Vivo CRISPR Screening Is Essential
Although in vitro screens offer rapid preliminary insights, they fail to fully recapitulate the complexity of in vivo biological systems, including the tumor microenvironment, immune surveillance, and pharmacokinetics. In vivo CRISPR library screening involves the introduction of gene-edited cells into living organisms, allowing for functional evaluation under authentic physiological conditions. This approach facilitates the identification of genes and pathways that are truly relevant in vivo, thereby generating target candidates with higher clinical translational value for therapeutic development.
What Targets Can Be Identified Using In Vivo CRISPR Screening?
1.Tumor Metastasis Targets
Tumor metastasis is the leading cause of cancer-related mortality. In vivo CRISPR screening enables the precise identification of key molecular mechanisms that drive metastatic progression. For example, in studies of breast cancer metastasis, researchers employed a genome-wide CRISPR activation library in an in vivo model and discovered that dysregulation of ribosomal protein expression and aberrant translational processes are critical factors promoting breast cancer dissemination.
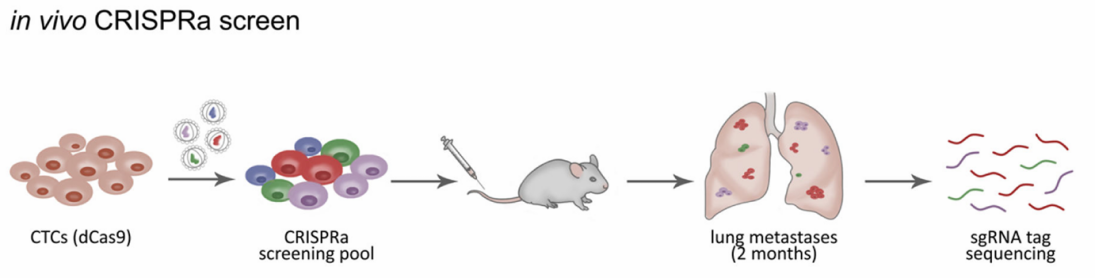
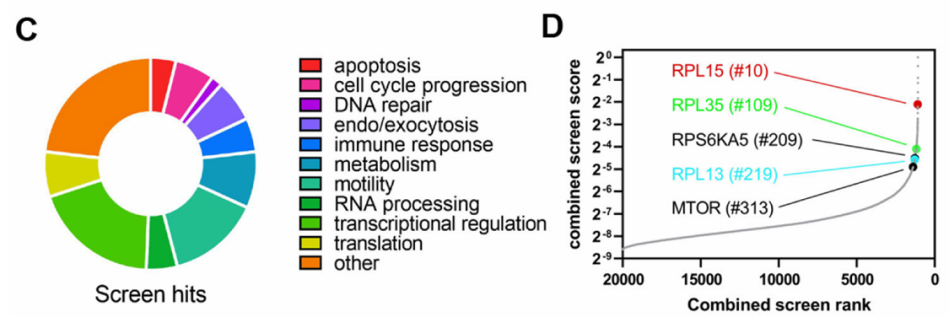
Figure 1. Genome-Wide CRISPR Activation Library Screening for Tumor Metastasis Targets.
Case Study: Identification of Metastasis Drivers Using a Genome-Wide CRISPR Activation Library
Researchers established a genome-wide CRISPR activation (CRISPRa) library cell line and intravenously injected two clones, Brx-82 and Brx-142, into NSG mice via the tail vein. Tumor cell migration and proliferation were monitored in vivo using live imaging. Two months post-injection, tumors had successfully metastasized to the lungs and exhibited robust growth. Subsequent next-generation sequencing (NGS) analysis of lung metastases revealed that overexpression of RPL15 markedly enhanced metastatic potential and selectively promoted the translation of other ribosomal proteins and cell cycle regulators.
2.Tumor Cell Therapeutic Targets
The proliferation of tumor cells in vivo is influenced by multiple factors, including the tumor microenvironment and inter-individual variability. By transitioning in vitro screening strategies to in vivo models, researchers can identify therapeutically relevant targets that are functionally effective within the physiological context. Typically, tumor-bearing mice are randomly assigned to experimental groups and subjected to specific interventions, enabling the discovery of targets that truly drive tumor growth and can be exploited for therapeutic purposes.
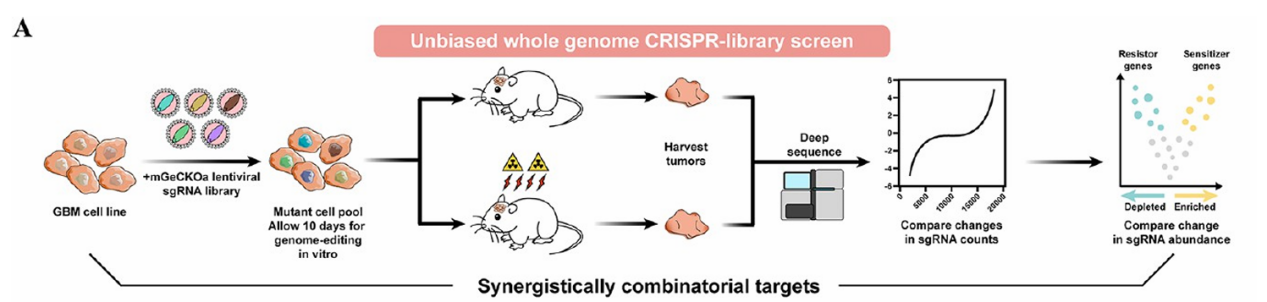
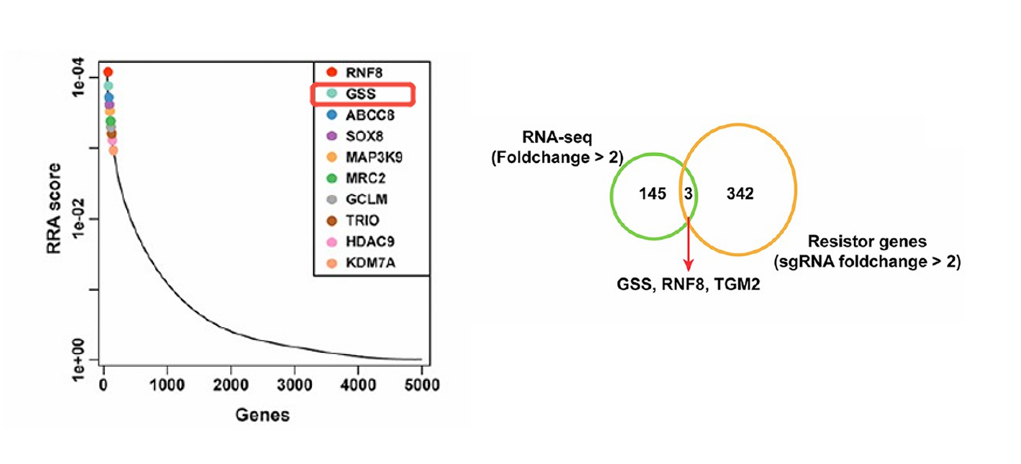
Figure 2. Genome-Wide CRISPR Knockout Library Screening for Tumor Therapeutic Targets.
Case Study: Identification of Tumor Therapeutic Targets Using a Genome-Wide CRISPR Knockout Library
Researchers constructed a genome-wide CRISPR knockout (CRISPRko) library in glioblastoma LN229 cells and implanted them into nude mice. Tumor-bearing mice were randomly assigned to experimental and control groups, with the experimental group receiving two weeks of irradiation. Subsequent next-generation sequencing (NGS) of tumor tissues, integrated with RNA-Seq data, revealed significant enrichment of the glutathione synthetase (GSS) gene in the screen. This finding suggests that GSS may modulate tumor cell radioresistance through regulation of ferroptosis pathways, highlighting a potential therapeutic target for enhancing radiotherapy efficacy.
3.Immune-Related Targets
Tumor immune evasion is a critical factor driving cancer progression and therapeutic failure. In vivo CRISPR library screening allows for the modeling of a physiologically relevant immune microenvironment, enabling the identification of key regulators that modulate tumor immune escape. This approach facilitates the discovery of targets that can enhance antitumor immunity and improve the efficacy of immunotherapies.
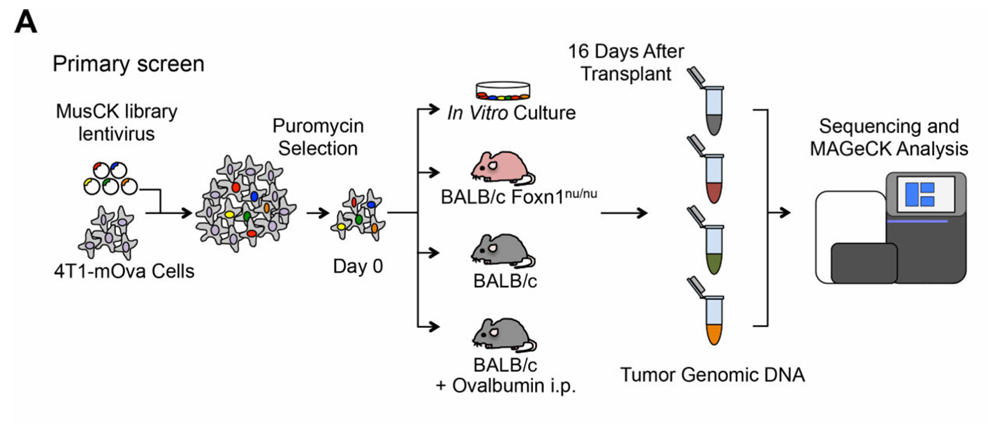
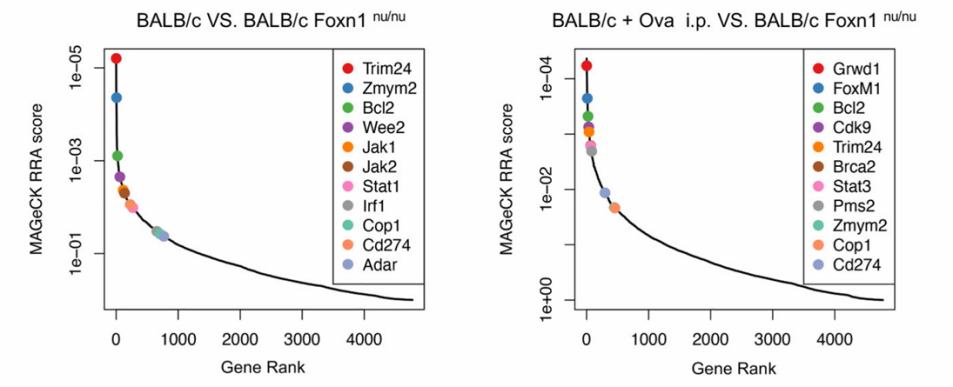
Figure 3. Customized CRISPR Library Screening for Immune-Related Targets.
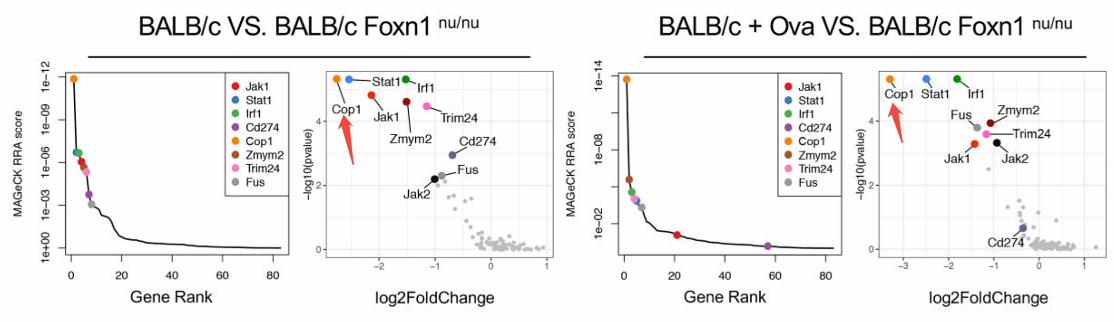
Figure 4. Secondary Screening Confirms COP1 as a Potential Immune-Modulatory Target.
Case Study: Identification of Immune-Modulatory Targets Using a Customized CRISPR Knockout Library
Researchers designed a CRISPR knockout library targeting 4,500 genes associated with tumorigenesis, progression, and immune regulation. The library was transduced into OVA-overexpressing 4T1-Ova cells and implanted into three mouse models: T-cell-deficient BALB/c nude mice, immunocompetent BALB/c mice, and BALB/c mice pre-immunized with OVA to enhance immune function. Approximately two weeks post-implantation, tumor tissues were collected for next-generation sequencing (NGS) analysis, identifying 79 significantly enriched genes. Secondary in vivo screening confirmed COP1 as the most enriched gene. Functional validation revealed that COP1 modulates chemokine secretion and macrophage infiltration within the tumor microenvironment, establishing it as a key target for improving immunotherapy efficacy in triple-negative breast cancer.
Advantages and Challenges of In Vivo CRISPR Screening
Advantages:
- Physiologically Relevant Modeling: In vivo screening more accurately replicates the complex physiological environment, producing results with higher clinical relevance.
- Comprehensive Functional Assessment: Enables systematic evaluation of gene function across multiple biological processes, including tumor initiation, metastasis, and immune evasion.
- Precise Target Identification: Provides reliable targets for drug development, effectively accelerating the preclinical discovery pipeline.
Challenges:
- Longer Experimental Timelines: Compared with in vitro screening, in vivo experiments require significantly longer durations and rely on specialized animal facilities and skilled personnel.
- Inter-Individual Variability: Biological differences among animals can reduce experimental reproducibility compared to in vitro models, necessitating increased sample sizes and experimental groups to ensure statistical reliability.
- Higher Experimental Costs: The resources required for in vivo screening are substantially greater than for in vitro approaches.
Ubigene Provides In Vivo CRISPR Screening Services
While in vivo CRISPR library screening can faithfully replicate physiological conditions and provide more reliable target identification, it requires specialized technical expertise. High-quality library construction, precise animal models, and professional data analysis are essential to ensure the reliability of screening results.
Ubigene possesses a comprehensive pre-experiment optimization and quality management system, offering one-stop, full-process in vivo CRISPR library screening services that seamlessly integrate each workflow stage. Additionally, our proprietary iScreenAnlys™ library analysis platform provides intelligent, free-of-charge data analysis. Researchers can choose from three major algorithms— Drug-Z、MAGeCK-RRA、MLE —and simply upload sequencing data to automatically obtain high-quality analysis results.
Contact us to learn more >>>Follow Ubigene to access more technical insights on CRISPR library screening and unlock the future of precision therapeutics together!
Reference
[1]Ebright RY, Lee S, Wittner BS, Niederhoffer KL, Nicholson BT, Bardia
A, Truesdell S, Wiley DF, Wesley B, Li S, Mai A, Aceto N, Vincent-Jordan
N, Szabolcs A, Chirn B, Kreuzer J, Comaills V, Kalinich M, Haas W, Ting
DT, Toner M, Vasudevan S, Haber DA, Maheswaran S, Micalizzi DS.
Deregulation of ribosomal protein expression and translation promotes
breast cancer metastasis. Science. 2020 Mar 27;367(6485):1468-1473.
[2]Liu X, Cao Z, Wang W, Zou C, Wang Y, Pan L, Jia B, Zhang K, Zhang W,
Li W, Hao Q, Zhang Y, Zhang W, Xue X, Lin W, Li M, Gu J. Engineered
Extracellular Vesicle-Delivered CRISPR/Cas9 for Radiotherapy
Sensitization of Glioblastoma. ACS Nano. 2023 Sep 12;17(17):16432-16447.
[3]Wang X, Tokheim C, Gu SS, Wang B, Tang Q, Li Y, Traugh N, Zeng Z,
Zhang Y, Li Z, Zhang B, Fu J, Xiao T, Li W, Meyer CA, Chu J, Jiang P,
Cejas P, Lim K, Long H, Brown M, Liu XS. In vivo CRISPR screens identify
the E3 ligase Cop1 as a modulator of macrophage infiltration and cancer
immunotherapy target. Cell. 2021 Oct 14;184(21):5357-5374.e22.


