What Does Luciferase Cell Do in Research?


What Does Luciferase Cell Do in Research?
What is a Luciferase Cell Line?
Luciferase cell lines are genetically modified cells that express luciferase proteins, which are enzymes that catalyze the oxidation of luciferin, resulting in bioluminescence. These cell lines are essential tools in molecular biology, allowing researchers to study gene expression and cellular processes in real-time. By using lentiviruses to introduce the luciferase gene into host cells, scientists can create stable luciferase-expressing cell lines that provide high sensitivity and specificity for various applications.
What is the Function of Luciferase?
The primary function of luciferase is to facilitate bioluminescence, enabling researchers to visualize and quantify biological processes without the interference of excitation light. This high signal-to-noise ratio makes luciferase an invaluable asset in numerous scientific fields, including cancer research, drug discovery, and gene expression studies. Ubigene offers high-quality luciferase cell lines that not only ensure stable expression of the luciferase gene but also provide strong specificity, high sensitivity, and excellent imaging quality.
Applications of Luc Cells
1.Application in vivo optical imaging experiment of living animals[1]
Bioluminescence imaging technology expressing biotinidase is the main method to study in vivo imaging of living animals. In vivo imaging technology can quickly and accurately screen and identify positive cell lines. The number of positive clones increased and the amount of luminescence increased, then the relationship between the number of positive cell lines and the luminous flux of the bioluminescent imaging system was discussed. The pMX-Luc2 plasmid was constructed by the molecular cloning method and cotransfection with pMD. G plasmid into 293T gag-pol cell line to obtain the retrovirus expressing Luc2 (Fig. 1). The retrovirus was used to infect mouse colon cancer CT26, human small cell lung cancer NCI-H446, human colon cancer HT-29, human ovarian cancer SKOV3, and human hepatoma SMMC-7721 cell lines to establish and screen Luc2 positive cell lines (Fig. 2). Different numbers of cell lines were inoculated in the culture dish, four nude mice were inoculated subcutaneously at different sites (dorsal or caudal vein) with 200μl different concentrations of SKOV3-Luc2 cell line. The luminous flux of the cell line inoculated in vitro and in vivo was detected by the bioluminescence imaging system. The results showed that the retroviral infection of Luc2 was a feasible method to establish the Luc2 positive cell line. The number of Luc2 positive cell line has a significant linear relationship with the luminous flux in the bioluminescent imaging system.
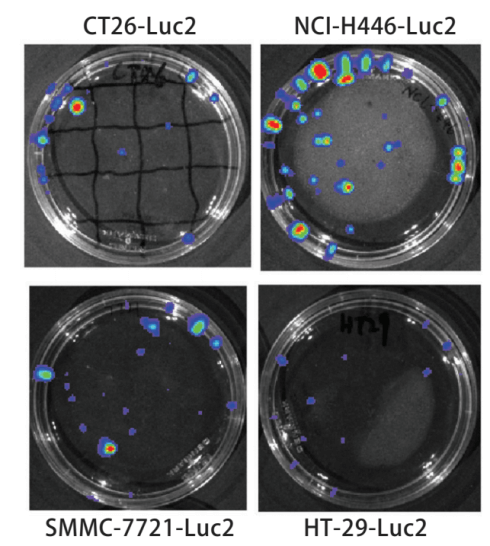
Figure 1. Bioluminescence test for Luc2-positive cell line
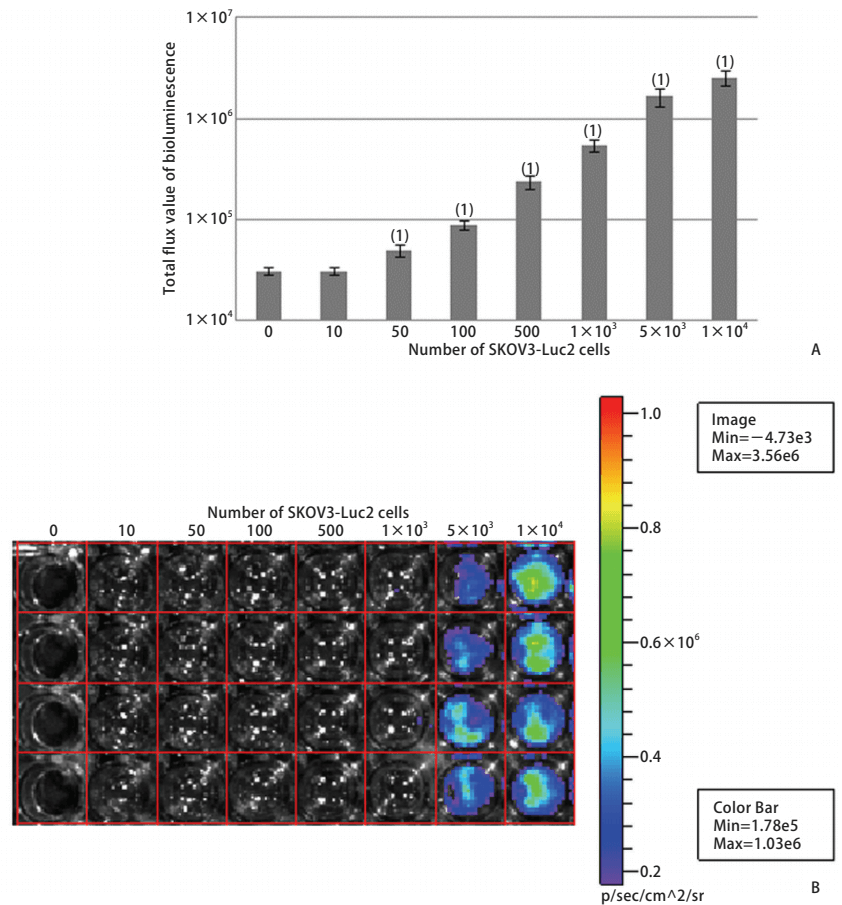
Figure 2. Bioluminescence luminous flux in SKOV3-Luc2 cell line (A), and Luminous flux diagram (B)
2.Verify the relationship between target gene and candidate gene[2]
To verify the relationship between FN1 (target gene) and MiR-96-5P (candidate gene), the sequence containing the binding site of FN1 and MiR-96-5P was cloned into luciferase reporter vector Psi-check2. Multiple comparisons showed that compared with the NC group, transfection of 100nM MiR-96-5P inhibitor significantly up-regulated the luciferase activity of cell line transfected with wild-type vector, but there was no significant change in the fluorescence activity of mutant vector (Fig. 3). he results of luciferase analysis showed that FN1 was the direct target gene of MiR-96-5P.
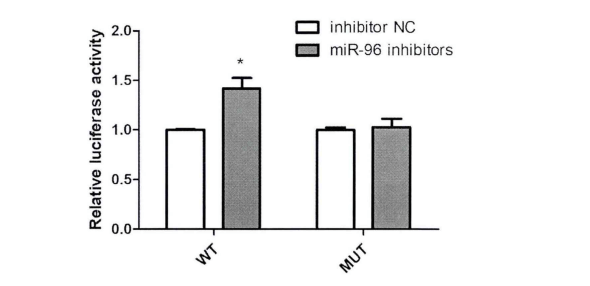
Figure 3. Fluorescence activity of HK2 cell line co transfected with MiR-96-5P inhibitor and WT/ mutant vector
3.Application of the pathological research[3]
The insulin 2 promoter was inserted into the PGL3-Basic vector to construct the plasmid with the luciferase reporter gene and transfection into islets β cell line MIN6, glucose induction was performed to detect insulin secretion by dual luciferase reporter gene system [Fig 4]. In the case of using PGI3-Basic blank plasmid instead of construction plasmid as blank control, the detection value of luciferase does not change with the induction of different concentrations of glucose, but remained at a low level (Fig. 4A), indicating that plasmid transfection had no effect on the test. Renilla luciferase was used as an internal reference to removing the differences between groups caused by the experimental operation. The results showed that (Fig. 4B and C) the expression value of Renilla in each group was basically the same, and the changing trend of each experimental group was the same after removing the influence of internal reference value.

Figure 4 . Effects of different concentrations of glucose on luciferase expression
4. Application in promoter activity analysis[4]
A single secretory luciferase reporter (SSLR) analysis method was developed and validated for promoter analysis. The method uses the early expression level of secretory luciferase associated with the test promoter as an internal standardized control for subsequent analysis of the same promoter (Fig. 5). SSLR analysis was compared with dual-luciferase reporter (DLR) analysis using HMGCR (3-hydroxy-3-methylglutamyl coenzyme A reductase) and LDLR (low-density lipoprotein receptor) promoter structures (Fig. 6). Similar results were obtained by both methods. Comparison of the reactions of HMGCR promoters in SSLR transient analysis is very advantageous compared to that of the same promoter in stable cell lines.
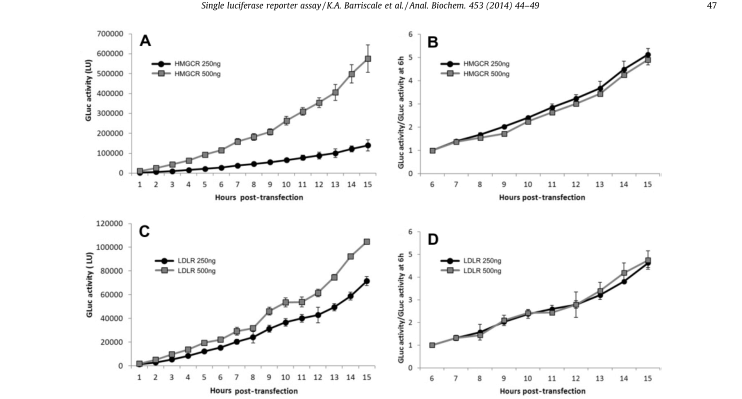
Figure 5. Luciferase activity/per hour after standardization.The data of secreted GLuc were standardized to the initial time point to correct the difference in plasmid transfection efficiency. HeLa cell line was transfected with 250 or 500 ng luciferase reporter for 6 to 9 hours. After removing the transfectant, the secretory luciferase activity was measured every hour. (A) The luciferase activity was measured after transfection with 250 or 500 ng pGluc-promHMGCR. (B) The luciferase activity of pGluc-promHMGCR was standardized for 6 hours. (C) The luciferase activity was measured after transfection with 250 or 500 ng pGluc-promLDLR. (D) The luciferase activity of pGluc-promLDLR was normalized to 6 hours.
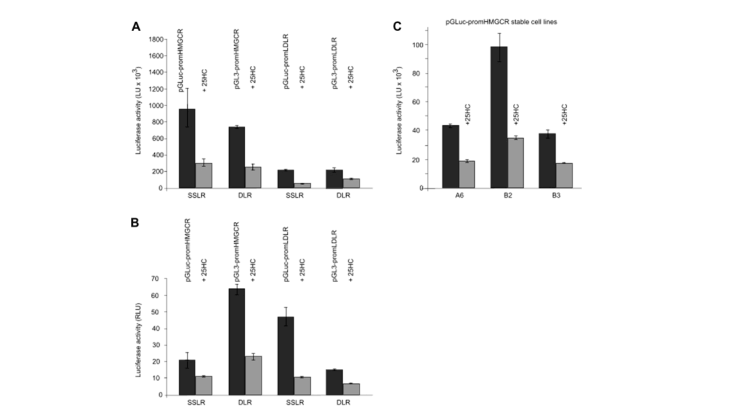
Figure 6. Comparison of SSLR analysis and DLR analysis. pGLuc-promHMGCR, pGL3-promHMGCR/pRL-TK, pGL3-promLDLR/pRLTK, or stable pGLuc-promHMGCR HeLa cell line were incubated under the condition of±25-OHC. For SSLR analysis samples, luciferase activity was measured 6 hours before 25-OHC treatment and 16 hours after 25-OHC treatment. (A) The promoter activity of HMGCR and LDLR of ±25-OHC was significantly lower than that of normal control. (B) The HMGCR and LDLR promoter activities of ±25-OHC were normalized to secrete luciferase at 6h and Renilla at DLR. (C) In stable pGLuc-promHMGCRHeLa cell lines A6, B2, and B3, the HMGCR promoter activity of ±25-OHC were detected.
Conclusion
luciferase reporter cell line shed light on different biological fields. Therefore, Ubigene generated a list of high-quality stable Luciferase cell lines. They can be directly applied to in vivo cell injection to detect tumorigenicity of the cell line. It also has the advantages of strong specificity and high sensitivity, high imaging quality, and accurate quantification of luminous intensity, which allows researchers to track their own cell line accurately .
Reference :
[1] Wang H J , Meng X T , Luan Q C , et al. Establishment of cell strains stably-transfected with luciferase gene mediated by retrovirus and their detection with bioluminescence imaging system[J]. Medical Journal of Chinese Peoples Liberation Army, 2012, 37(5).
[2] Wang W , Jia Y J , Yang Y L , et al. LncRNA GAS5 exacerbates renal tubular epithelial fibrosis by acting as a competing endogenous RNA of miR-96-5p[J].Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie, 2020, 121:109411.
[3] 康宇佳, 蒙明慧, 吕忠显. PGL3-insulin Luciferase质粒的构建与功能鉴定[J]. 动物医学进展, 2012, 33(004):10-15.
[4] Kathy A, et al.A single secreted luciferase-based gene reporter assay[J],Analytical Biochemistry,2014,453 (2014) 44–49.


