Identification of Critical Genes Driving Mouse Tumor Metastasis via CRISPR Screening


Identification of Critical Genes Driving Mouse Tumor Metastasis via CRISPR Screening
Background
Cancer metastasis is a highly complex process involving the coordinated regulation of multiple genes and is characterized by dynamic changes and pronounced heterogeneity. Tumor metastasis is not only governed by intrinsic genetic programs of cancer cells but is also influenced by the complex physiological microenvironment in vivo, which cannot be faithfully recapitulated by simple, stable in vitro culture systems. Traditional approaches to cancer target research typically rely on existing literature, protein function studies, or clinical data to validate the function of a single or a few candidate genes. While this strategy allows for in-depth investigation of known gene-disease relationships and mechanisms, it has significant limitations: it is low-throughput, requires hypothesis-driven experiments, and is less suitable for discovering unknown pathways or novel potential targets.In recent years, CRISPR library screening has emerged as a high-throughput, systematic approach for functional genomic analysis and has been increasingly applied in cancer-targeted drug discovery. Compared with conventional methods, CRISPR library screening offers several advantages:
- - High throughput:enabling genome-wide or large-scale gene coverage in a single experiment;
- - High efficiency:directly linking phenotypes to functional genes;
- - Low off-target effects:allowing precise editing of target genes while minimizing non-specific effects.
By integrating CRISPR library screening with traditional research strategies, it is possible to maintain in-depth studies of known targets while simultaneously identifying novel key genes through phenotype-based screening, thereby accelerating target discovery and functional validation. In this context, Zhang et al., 2015, in their study “Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis”, for the first time applied a genome-wide CRISPR screen in an in vivo mouse model to systematically identify genes that regulate tumor growth and metastasis within a physiological microenvironment, providing a new experimental framework for investigating tumor metastasis mechanisms in vivo.
CRISPR Library Reveals Core Genes for Mouse Tumor Metastasis
1. Cell Model
Mouse non-small cell lung cancer cells (KrasG12D-/+;p53 -/- ;Dicer1+/-, abbreviated as KPD cells) were used. KPD cells were first transduced with Cas9-GFP lentivirus to generate a stable Cas9-GFP KPD cell line.
2. CRISPR Library Design
A mouse genome-wide sgRNA library (mGeCKOa) was employed, comprising 67,405 sgRNAs targeting 20,611 protein-coding genes and 1,175 pre-miRNAs. In addition, 1,000 non-targeting sgRNAs were included as controls, providing a basis for high-throughput functional screening.
3. Cell pool generation and Positive Selection
- - Lentiviral Infection:Three independent replicate experiments were performed, with each achieving a coverage >400× and an average MOI of 0.4 ± 0.02.
- - Positive Cell Selection:Infected cells were cultured with 2 μg/mL puromycin for 7 days to enrich for successfully transduced cells.
4. CRISPR In Vivo Screening Strategy
The lentivirus-infected KPD cells were subcutaneously injected into the right flank of nude mice. For each of the three replicate infections, cells were injected into four mice: one mouse for early-stage primary tumor sequencing and the remaining three for late-stage primary tumor and distant metastatic lesion sequencing. Wild-type (uninfected) cells were injected as controls.
Six weeks post-injection, mice were sacrificed and tumor formation and metastasis were evaluated:
① Primary tumors at injection site: collected to identify genes essential for primary tumor growth.
② Distant metastatic lesions in various organs: collected to identify genes required for metastatic colonization.
5. CRISPR Screening Logic
A combination of negative and positive selection was employed by comparing sgRNA representation across the library cells, primary tumors, and metastatic lesions:
① Depleted sgRNAs in primary tumors or metastatic lesions → corresponding genes are essential for tumor growth and metastasis formation.
② Enriched sgRNAs in primary tumors or metastatic lesions → corresponding genes suppress primary tumor growth and metastasis formation.
Key Findings
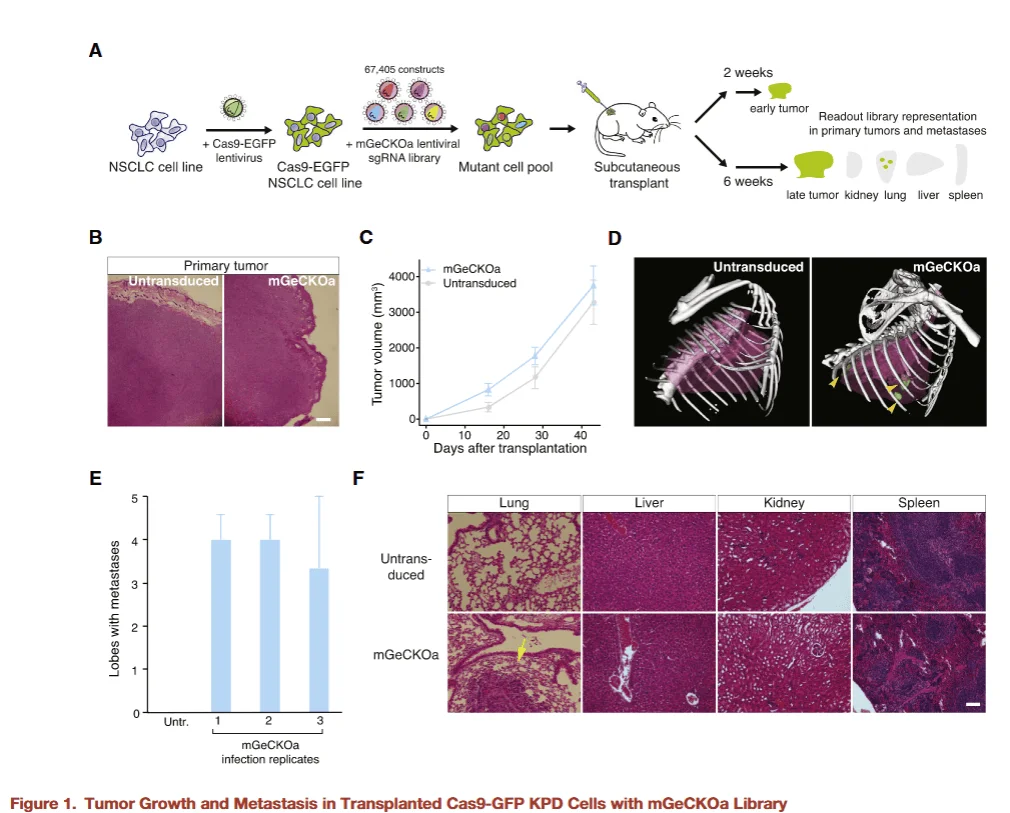
Figure 1
The researchers first generated mouse genome-wide knockout (KO) library cells and subcutaneously injected them, alongside wild-type control cells, into the flanks of mice to assess the impact of gene loss on primary tumor formation and metastatic potential. The experimental results were as follows:
- - Primary tumor formation:Both the library-injected group and the control group successfully developed primary tumors at the injection site, indicating that the gene knockouts did not impair local tumor growth.
- - Distant metastasis:Metastatic lesions in the lungs were detected only in mice injected with library cells, suggesting that the genome-wide KO library contains genes whose loss promotes tumor metastasis.
This finding demonstrates for the first time in an animal model that a genome-wide CRISPR KO library can systematically identify potential genes closely associated with tumor metastasis, providing a foundation for further mechanistic studies of metastatic progression.
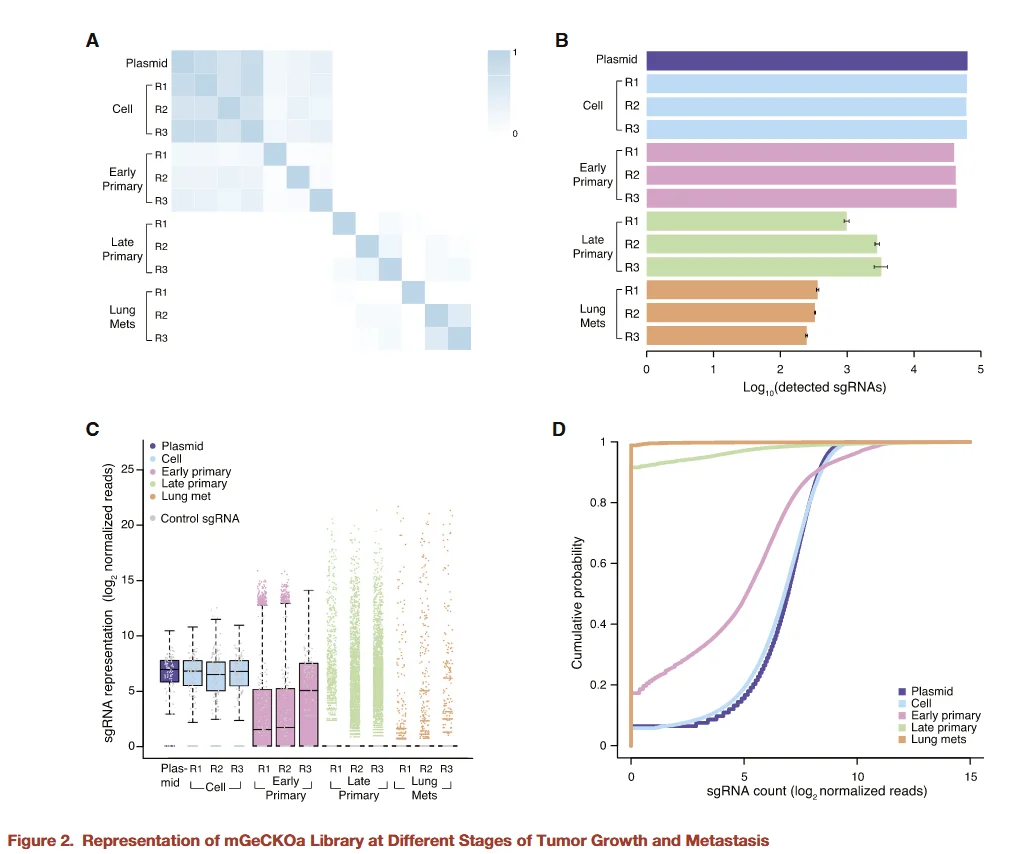
Figure 2
To further investigate the dynamic changes in sgRNA representation during tumor initiation and metastasis, the researchers sequenced sgRNAs from library plasmids, library-infected cells, early-stage primary tumors, late-stage primary tumors, and lung metastases. The results were as follows:
- - Library plasmids vs. library cells:sgRNA abundances were largely consistent, indicating that viral infection and cell expansion did not introduce significant bias.
- - During tumor progression and metastasis:As tumors progressed from early-stage primary tumors to late-stage primary tumors and finally to lung metastases, the abundance of certain sgRNAs gradually decreased (early primary tumor > late primary tumor > lung metastasis). This suggests that, during tumor initiation, progression, and metastasis, cells harboring specific gene knockouts experienced selective pressures, leading to the enrichment or depletion of the corresponding sgRNAs.
These dynamic changes provide an experimental basis for identifying genes essential for tumor growth and metastasis and highlight the critical role of the in vivo microenvironment in shaping the functional selection of tumor cell genes.
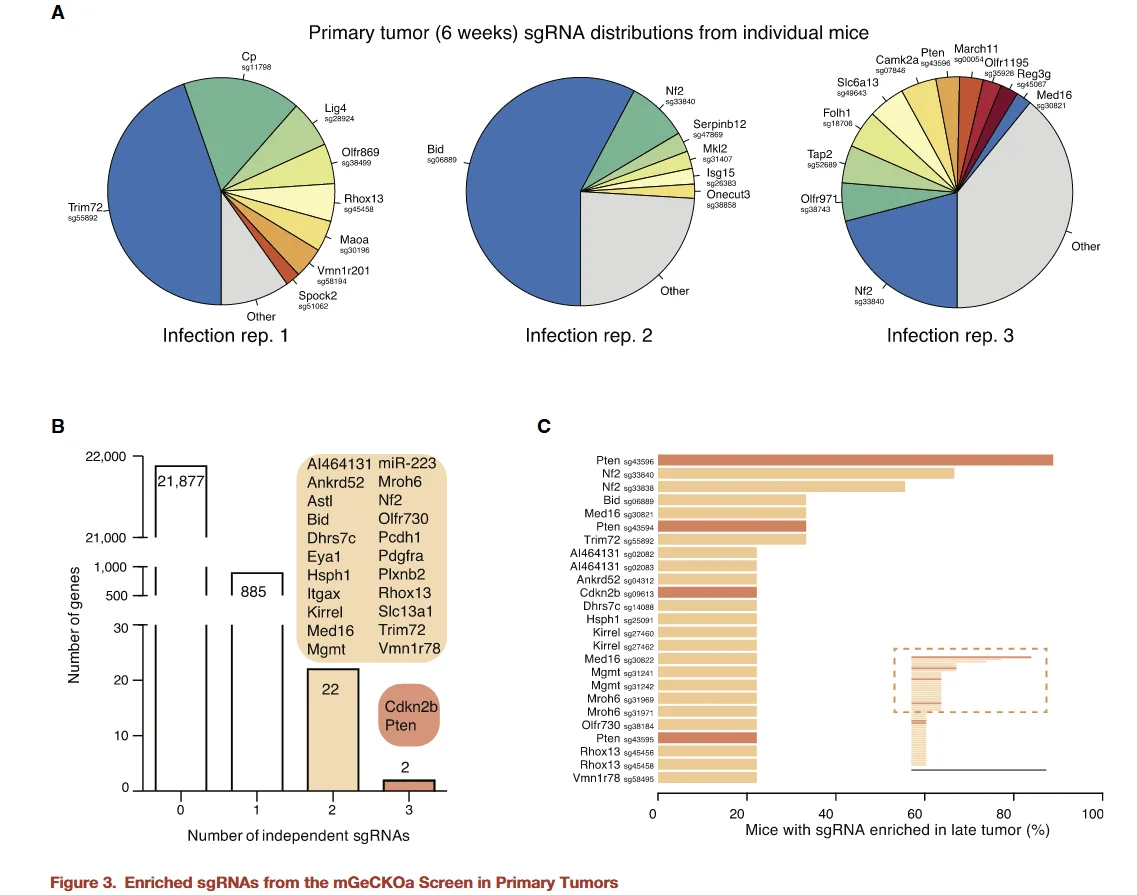
Figure 3
Following the acquisition of sequencing data from mouse primary tumors, the researchers first analyzed sgRNA representation within the primary tumors. Using non-targeting sgRNAs as controls and applying a threshold of FDR < 0.2%, sgRNAs from the primary tumors of three mice were statistically evaluated. The results were as follows:
- - A total of 935 sgRNAswere significantly enriched in at least one mouse, targeting 909 genes.
- - Functional features:Many of these genes are associated with apoptosis, suggesting that they may play critical roles in primary tumor growth and metastasis.
- - Highly consistent genes:Among them, 24 genes had enriched sgRNAs in two or more mice, indicating high reproducibility and potential biological significance.
This analysis provides a foundation for the subsequent functional validation of key genes in tumor metastasis and highlights the potential central role of apoptosis-related genes in regulating metastatic progression.
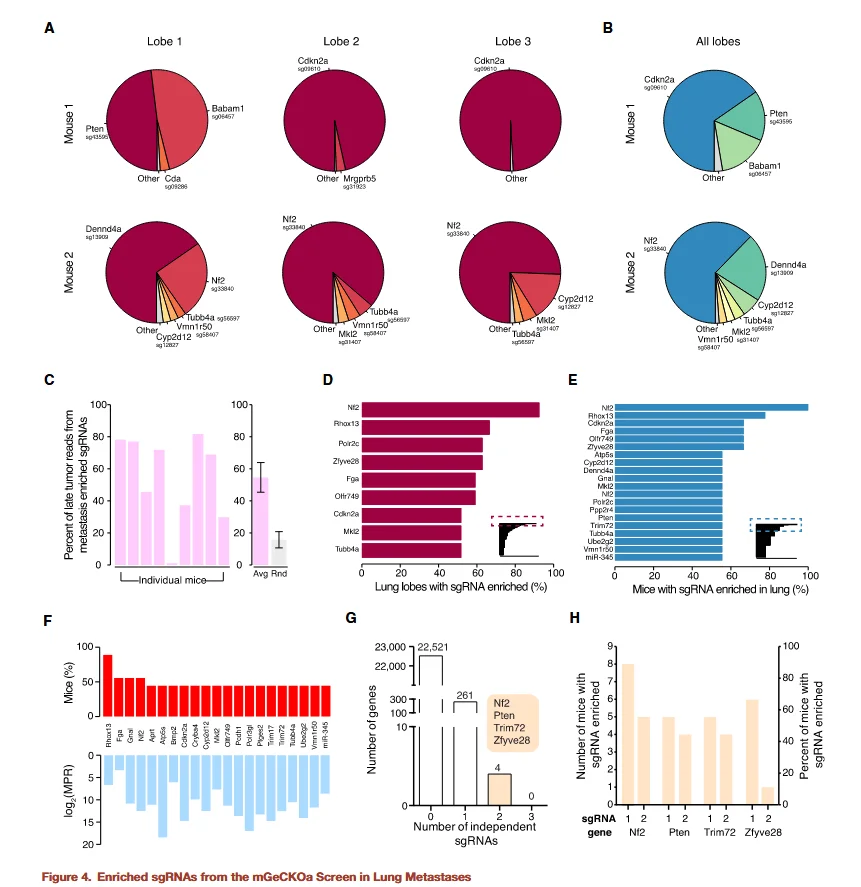
Figure 4
The researchers further analyzed sgRNA representation in metastatic lesions, again using non-targeting sgRNAs as controls and applying a threshold of FDR < 0.2%. The results were as follows:
- - Total enriched sgRNAs:A total of 147 sgRNAs were significantly enriched in at least one lung lobe, with 105 sgRNAs detected in the lung lobes of at least one mouse.
- - Key genes:The enriched sgRNAs targeted genes including Nf2, Pten, Cdkn2a, Trim72, Fga, Zfyve28, Rhox13, Babam1, miR-345, and miR-152.
- - Specificity analysis:For several genes, sgRNA abundance in metastatic lesions was higher than in primary tumors (MPR > 1), suggesting that these genes are specifically required for the metastatic process rather than for simple cell proliferation.
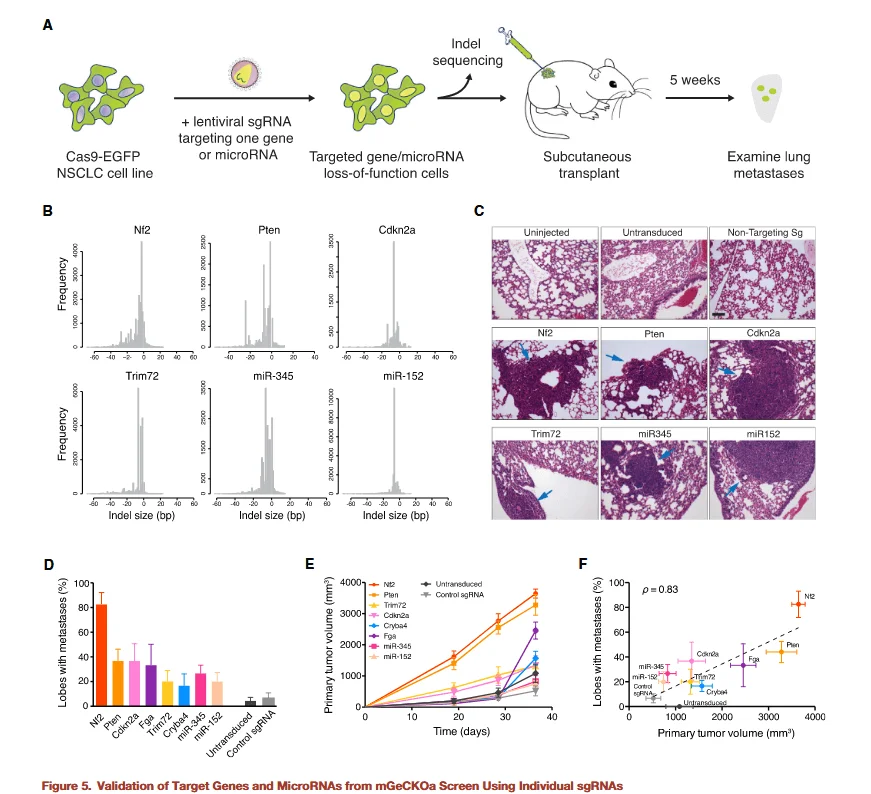
Figure 5
To validate the functional roles of the identified candidate genes, the researchers generated single-gene knockout (KO) cell lines and injected them into mice to assess lung metastasis formation. Wild-type cells and non-targeting sgRNA KO cells were used as controls.
The experimental results were as follows:
- - Enhanced metastasis:Knockout of all candidate genes significantly promoted the formation of metastatic lesions in the lungs.
- - Dual effects on tumor growth and metastasis:Some gene knockouts not only enhanced primary tumor proliferation but also increased metastatic colonization.
- - Consistency with library screening predictions:These observations were highly consistent with the predictions from the genome-wide CRISPR library screening and bioinformatic analyses.
This validation demonstrates the reliability and reproducibility of the CRISPR library screening results and further confirms the critical roles of the candidate genes in tumor metastasis.
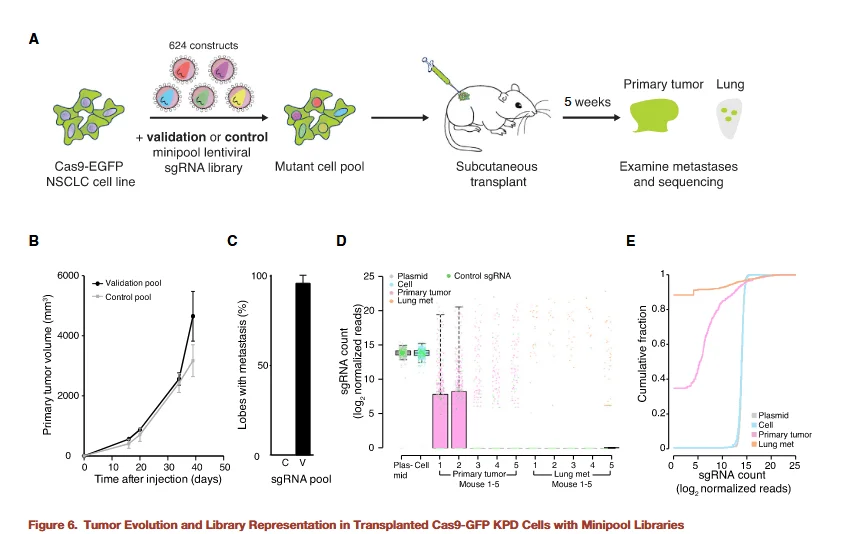
Figure 6
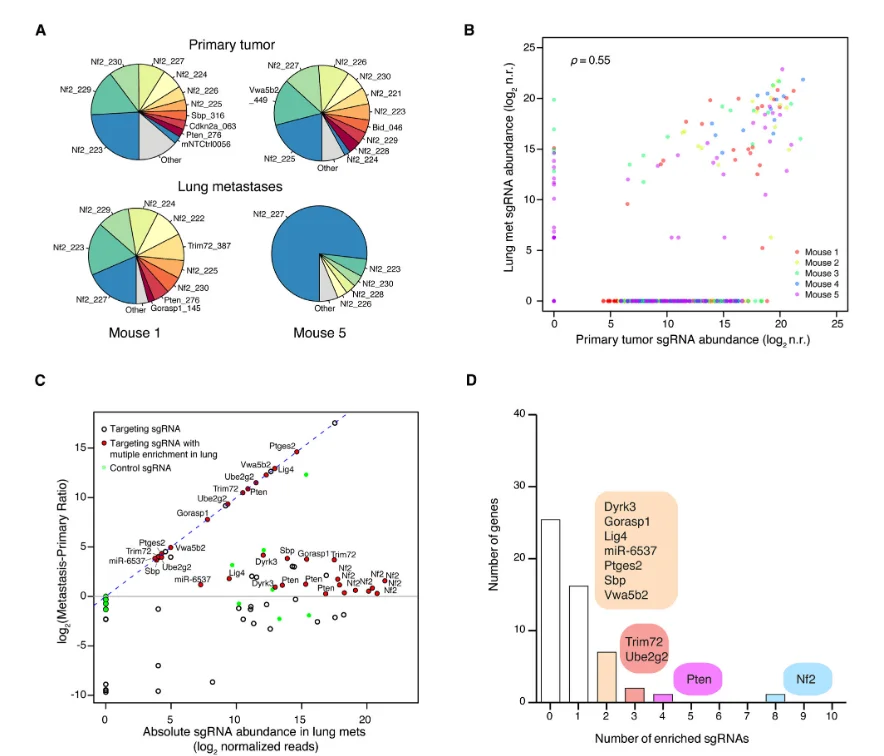
Figure 7
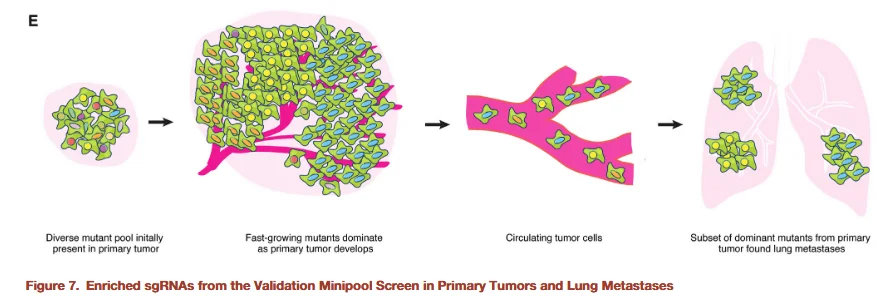
Figure 8
To further validate the identified candidate genes, the researchers constructed a small-scale sgRNA library designed to increase screening depth while reducing off-target effects and background noise associated with large genome-wide libraries. This mini-library targeted only 53 candidate genes, with multiple sgRNAs per gene to enhance the reliability of validation. Experiments using this small library again demonstrated that gene knockouts promoting primary tumor proliferation also enhanced metastatic lesion formation, further confirming the accuracy of the genome-wide screening results.
To evaluate the clinical relevance of these findings, the corresponding human homologs of the candidate genes identified in mice were queried in the TCGA database. The analysis revealed that these genes were predominantly downregulated in metastatic lesions of patients with non-small cell lung cancer (NSCLC, 61%–75%), suggesting that the screening results obtained from the mouse model are clinically informative and may provide valuable insights for studying human cancer metastasis and identifying potential therapeutic targets.
Clinical Significance
This study employed a genome-wide CRISPR library to perform high-throughput screening, comparing sgRNA enrichment between primary tumors and metastatic lesions to identify multiple candidate genes, which were subsequently validated through single-gene functional assays. The researchers then constructed a small-scale sgRNA library for further validation of these candidates, and finally compared the experimental results with human gene expression data from existing databases, forming a complete experimental and analytical framework.
This approach not only provides a direct reference for subsequent functional studies or the development of targeted therapies, but also serves as a model framework for systematic functional gene screening in other diseases or physiological processes.
Key Highlights
- - Innovative methodology:This study demonstrates for the first time the feasibility of performing genome-wide CRISPR screening in vivo, providing a novel technical framework for tumor research.
- - Potential metastasis-related targets:Genes such as Nf2, Pten, Cdkn2a, Trim72, Fga, miR-345, and miR-152 were identified, whose loss-of-function accelerates the formation of metastatic lesions.
- - Clinical translational value:These genes may serve as prognostic biomarkers for cancer patients and represent potential therapeutic targets to inhibit tumor metastasis.
Summary of Core Concept
By employing CRISPR-Cas9 technology for genome-wide loss-of-function screening, this study systematically identified genes that play key roles in tumor initiation, progression, and metastasis within an in vivo environment. CRISPR library screening offers advantages such as high throughput, low off-target effects, and unbiased discovery, enabling the systematic identification of all potential functional genes. By integrating CRISPR library screening with in vivo experiments, this study precisely uncovered genes associated with tumor growth and metastasis in a physiological context, providing novel tumor-related targets and a replicable experimental framework for subsequent research. These findings offer robust support for cancer mechanism studies and drug development.
Ubigene CRISPR Screening Services
To empower researchers in efficiently identifying functional genes in tumor and immunology research, Ubigene offers a comprehensive CRISPR screening service, covering in vitro screening, in vivo screening, and full data analysis workflows. Leveraging the iScreenAnlys™ CRISPR library analysis platform, researchers can perform high-throughput, low off-target, and fully visualized data processing and target analysis.From experimental design to result interpretation, Ubigene provides full support for universities, research institutes, and pharmaceutical R&D organizations, making CRISPR library screening efficient, controllable, and reproducible, thereby accelerating scientific breakthroughs and target discovery.
Contact us now to start your high-efficiency target screening journey!>>
Reference
[Chen S, Sanjana NE, Zheng K, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160(6):1246-1260. doi:10.1016/j.cell.2015.02.038]



