Multiplex CRISPRa Combined with Single-Cell Sequencing: Deciphering Cell-Type-Specific Gene Regulatory Elements


Traditional CRISPR is well-known as a "gene scissor", capable of precisely cutting DNA. In contrast, CRISPRa (CRISPR activation) functions more like a “gene volume control,” upregulating gene expression without altering the DNA sequence itself. CRISPRa employs a catalytically inactive Cas9 (dCas9) fused to transcriptional activator domains, enabling reversible and finely tuned gene regulation with significant potential for both research and therapeutic applications.
However, conventional CRISPRa screens face a critical limitation: they are often unable to assess cell-type-specific regulatory effects. This is particularly important for minimizing off-target effects in gene therapy. A study published in Nature Communications in September 2024 proposed a novel strategy to address this challenge. By integrating multiplex CRISPRa screening with single-cell RNA sequencing, the researchers established a multiplex single-cell CRISPRa screening platform capable of identifying cell-type-specific regulatory elements, offering unprecedented resolution for dissecting gene regulation across diverse cellular contexts.
Research Background
The human genome contains millions of candidate cis-regulatory elements (cCREs), yet only a small fraction have been functionally validated and reliably linked to their target genes. Previously, the authors' team, in collaboration with other researchers, employed CRISPR interference (CRISPRi) combined with single-cell RNA sequencing to perform large-scale validation of distal cCREs and successfully associated them with their regulatory targets. However, to date, CRISPRa has not been widely applied in non-coding element screens, and additional data are needed to establish practical guidelines for its use. Most studies in this field have focused primarily on assessing the necessity of candidate regulatory elements, with only a few attempting to verify their sufficiency in the endogenous context.
Traditional methods face significant challenges in matching regulatory elements with their target genes at scale. Low infection efficiency and the limitations of single-guide RNA (sgRNA) design prevent comprehensive capture of gene expression changes. On one hand, conventional CRISPRa libraries rely on low-efficiency delivery to achieve single sgRNA integration per cell, which is insufficient for efficiently screening the millions of cCREs against approximately 20,000 regulated genes in the human genome. On the other hand, traditional CRISPRa approaches typically focus on the regulation of individual genes and subsequent expression profiling of specific targets, making it difficult to systematically understand the broader gene expression changes induced by perturbations.
A Multiplexed Single-Cell CRISPRa Screening Strategy
The authors developed an innovative screening framework (Fig. 1) that overcomes key limitations of conventional pooled CRISPRa libraries. The core elements of this design include:
- Randomized gRNA Combinations: Using the piggyBac transposon system, multiple gRNAs are stochastically integrated into individual cells, enabling multiplexed perturbations while substantially reducing the required cell input. These randomized gRNA combinations drive stronger transcriptional perturbations, markedly improve the sensitivity for detecting gene expression changes, and greatly diminish the number of cells needed for downstream analyses.
- Single-Cell RNA Sequencing (scRNA-seq): Instead of gene-specific readouts, the authors employ high-resolution 10x Genomics scRNA-seq, enabling comprehensive transcriptome-wide measurement in a single experiment without the need to predefine target genes.
- Computational Assignment: Differential expression analysis is then used to accurately map each gRNA to its regulatory effects on cognate target genes.
In summary, this strategy offers exceptional throughput—allowing large-scale, combinatorial interrogation of regulatory elements within a single experiment—while significantly reducing both time and resource consumption.
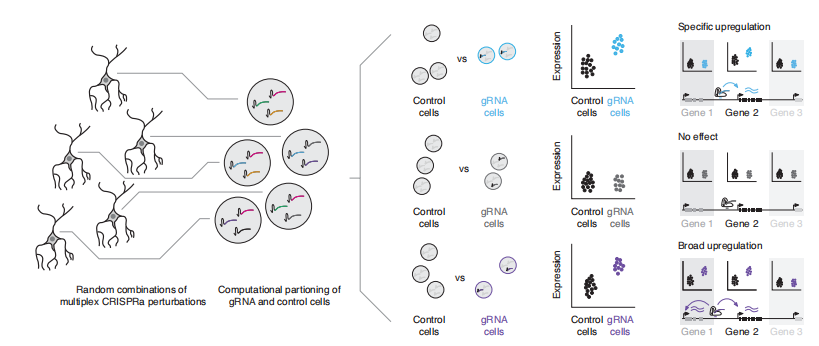
Figure 1. Schematic Overview of the Multiplexed Single-Cell CRISPRa Screening Strategy
Experimental Design
-
Cell Lines
The study employed two representative cellular models—K562 chronic myelogenous leukemia cells and human induced pluripotent stem cell (iPSC)-derived excitatory neurons—to evaluate whether candidate regulatory elements exhibit cell type-specific activity. -
Construction and Validation of dCas9 Activation Complexes
Because no consensus has been reached regarding the most effective CRISPRa activation complex for broad and scalable targeting of cCREs, the authors benchmarked two widely used dCas9-based activators: the VP64 activator (comprising four tandem VP16 effector domains) and the VPR activator (a tripartite fusion of VP64, p65, and Rta effectors). To rigorously assess CRISPRa activity, a CMV promoter-driven tdTomato reporter system was used, enabling quantitative evaluation based on tdTomato expression. -
gRNA Library Design and Construction
Guide RNA design represents a critical step for achieving reliable functional validation. The workflow was structured to ensure effective identification of gRNAs associated with cell type–specific regulatory elements. The design process included:- Selection of Positive Control gRNAs: Thirty gRNAs targeting regions near transcription start sites (TSSs) were randomly drawn from the validated hCRISPRa-v2 library described by Horlbeck et al.
- Selection of Negative Control gRNAs: Fifty non-targeting control (NTC) gRNAs were similarly chosen from the hCRISPRa-v2 library.
- Design of Candidate Promoter-Targeting gRNAs: A total of 313 gRNAs were designed to target 50 candidate TSSs associated with nine neurodevelopmental disorder (NDD) risk genes (TCF4, FOXP1, SCN2A, CHD8, BCL11A, TBR1, SHANK3, SYNGAP1, and ANK2).
- Design of Candidate Enhancer-Targeting gRNAs: Fifty gRNAs were designed to target 25 enhancer loci previously validated as “active” by CRISPRi experiments from the authors' group. An additional fifty gRNAs targeted 25 enhancer loci previously marked as “inactive” in CRISPRi assays but predicted to function as strong enhancers in K562 cells.
Results
Through the pooled screening performed in K562 cells, the study successfully identified multiple promoter- and enhancer-targeting gRNAs capable of robustly upregulating gene expression, yielding a total of 59 activation hits (Fig. 2). These hits span diverse categories, including promoter-targeting gRNAs for genes such as FOXP1 and CHD8 , as well as enhancer-targeting gRNAs associated with genes like ANK2 . Notably, the activated gRNAs exhibited high target specificity and were generally located in close genomic proximity to their respective genes, providing valuable insight into the regulatory architecture governing transcription in K562 cells.
In addition, the study uncovered enhancers that, when targeted individually, were sufficient to upregulate their associated genes. This observation challenges the traditional view that enhancer activity must operate strictly in concert with promoter elements, and highlights the diversity and complexity of gene regulatory mechanisms.
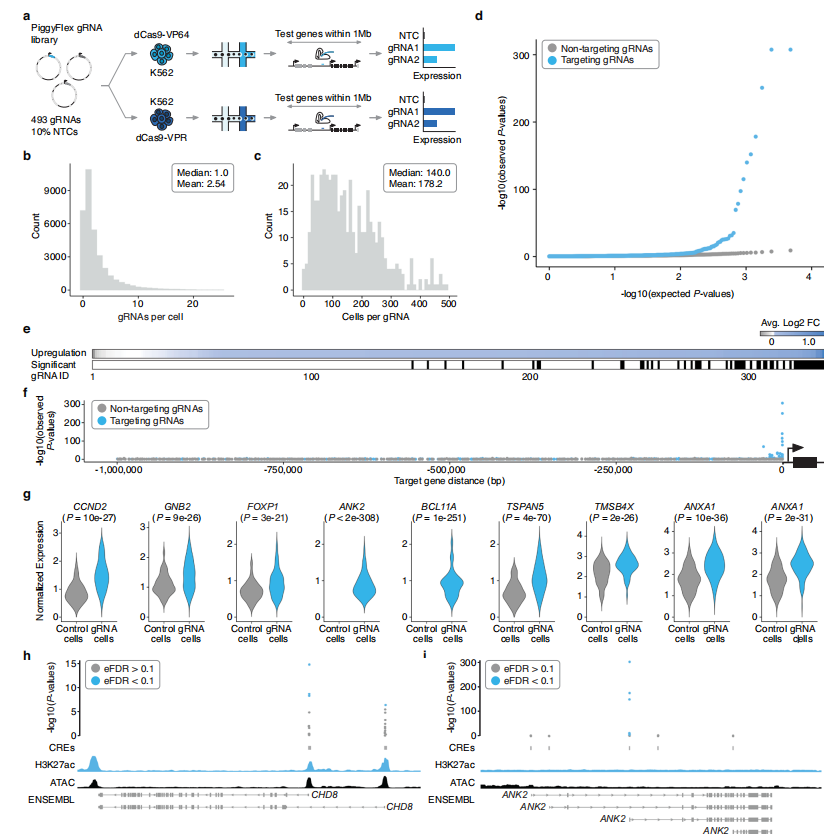
Figure 2. Multiplexed Single-Cell CRISPRa Screening of Regulatory Elements in K562 Cells
In the screen performed in iPSC-derived neurons, a stringent empirical FDR threshold of 0.1 yielded 17 hit gRNAs. All of these corresponded either to TSS-targeting positive controls or to candidate promoters of ASD/NDD-associated neurodevelopmental risk genes such as TCF4, FOXP1, and TBR1 (Fig. 3). The results demonstrate strong target specificity in neurons—for example, the TBR1-targeting gRNA selectively upregulated TBR1 expression in neuronal cells but showed no such effect in K562 cells. This finding highlights the exceptional sensitivity of the screening framework for identifying cell type-specific regulatory elements and provides key insights for dissecting gene regulatory mechanisms underlying neuronal development and function.

Figure 3. Multiplexed Single-Cell CRISPRa Screening of Regulatory Elements in Postmitotic iPSC-Derived Neurons
To further validate the robustness of the screening results, the researchers reanalyzed the dataset using the SCEPTRE statistical framework. SCEPTRE identified an even larger number of hits, with strong concordance relative to the original analytical pipeline, thereby providing compelling evidence for the stability and reliability of the findings (Fig. 4).
In addition, the team performed carefully designed, independent single-target perturbation experiments to validate eight representative hit gRNAs. Sequencing results confirmed that most gRNAs upregulated their intended target genes in the expected cell type. The outcomes of these individual validations showed strong agreement with the multiplexed single-cell screening results (Pearson's correlation coefficient = 0.83) , offering solid experimental support and further reinforcing the credibility of the study's conclusions.
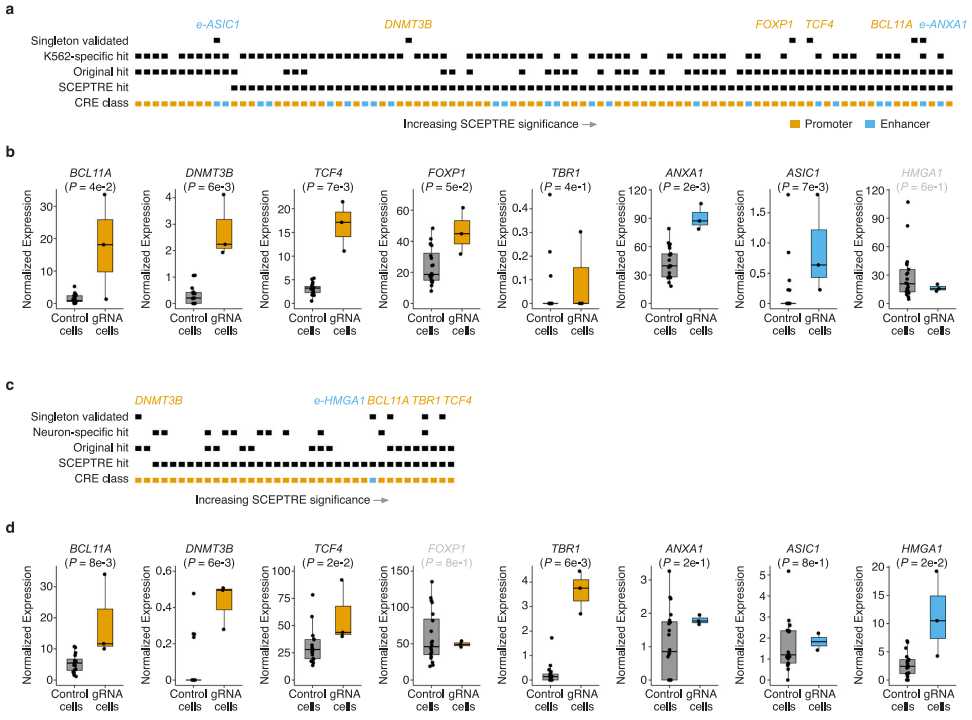
Figure 4. Covariate-Aware Reanalysis and Single-Perturbation Validation Supporting and Extending the Screening Results
Taken together, these findings refine and further extend the multiplexed single-cell CRISPRa screening framework, enabling the successful identification of functional and cell type-specific endogenous CREs that regulate haploinsufficient genes in both K562 cells and iPSC-derived excitatory neurons.
In summary, the multiplexed single-cell CRISPRa platform demonstrates a powerful capacity to identify CRISPRa-responsive regulatory elements—and their corresponding gRNAs—with high target and cell type specificity. Moreover, it facilitates the discovery of CREs that operate uniquely across different cellular contexts, offering a scalable and high-resolution approach for elucidating complex gene regulatory landscapes.
Prospects and Applications
This study offers broad application potential, spanning fundamental biological research to translational and therapeutic development.In basic research , the framework enables construction of comprehensive, cell type-specific enhancer-gene regulatory maps, providing powerful insights into the mechanisms governing gene expression across biological processes.
In disease-focused studies, the method can be used to systematically characterize the functional impact of disease-associated noncoding variants, helping to elucidate the mechanisms underlying risk loci uncovered by genome-wide association studies (GWAS).From a clinical and therapeutic perspective, the platform holds promise for large-scale identification of gRNAs and cell type-specific CREs relevant to haploinsufficiency-related disorders. This creates a technical foundation—and a conceptual roadmap—for developing cell type-specific gene regulatory therapies.
Looking ahead, future work may further expand the scale and breadth of the screens by incorporating additional cell types and developmental stages, ultimately enabling a more complete and fine-grained understanding of human gene regulatory networks.
Ubigene CRISPR Screen Services
Leveraging our proprietary CRISPR-iScreen™ technology, Ubigene has established a versatile CRISPR screening platform encompassing CRISPR-KO、CRISPRa、CRISPRi approaches. We offer end-to-end, one-stop CRISPR screening services, with seamless integration across all workflow stages to efficiently achieve target discovery.Our CRISPR screening services cover both in vitro and in vivo applications:
- In vitro screening provides diverse phenotypic analysis options, enabling customized experimental designs to meet specific research needs.
- In vivo screening precisely recapitulates the physiological microenvironment and is compatible with a variety of mouse models, offering reliable data that closely reflect true biological conditions and support robust target validation.
Reference
Chardon FM, McDiarmid TA, Page NF, Daza RM, Martin BK, Domcke S, Regalado SG, Lalanne JB, Calderon D, Li X, Starita LM, Sanders SJ, Ahituv N, Shendure J. Multiplex, single-cell CRISPRa screening for cell type specific regulatory elements. Nat Commun. 2024 Sep 18;15(1):8209. doi: 10.1038/s41467-024-52490-4. PMID: 39294132; PMCID: PMC11411074.


