Prime Editing: Revolutionary Breakthrough in Precise Gene Therapy


Prime Editing: Revolutionary Breakthrough in Precise Gene Therapy

Nowadays, many are faced with the need of constructing a point mutation cell line at the gene of interest (GOI). Luckily, scientists have developed a variety of editing approaches, such as Ribonucleoprotein(RNP) delivery (direct and with high efficacy, electroporation devices needed), Base Editors (BE, for mapped swapping of a single basepair) and Plasmid Resistance Screening (Selecting resistant cell colonies to improve edit rate). Each of these methodologies is like a tool in a toolbox, and researchers need to select the appropriate one based on their respective application scenarios and technical advantages. Recently joining this array is Prime Editing (PE), which we would like to introduce and demonstrate why Prime Editing has become popular and well practiced in the past few years.
I. What is Prime Editing?
Prime Editing was first proposed by David Liu’s team at Havard University in 2019, as a precise gene editing method featuring being“breakage-less and programmable”. To be differentiated from traditional CRISPR-Cas9 editing systems, which relies on introducing a double-strand breakage (DSB), Prime Editing facilitates a “search and replace” mechanism of action through a protein fusion of the Cas9 nickase (nCas9) and Reverse Transcriptase (RT). This significantly reduces the off-target rate and the risk of chromosomal rearrangements. It also expands the scope of gene editing, highlighting the importance of prime editing in enabling safer and more flexible genomic modifications.
Advantages of Prime Editing
1. Precise, inspecific single base swapping (not limited to A>G or C>T, like that in BE)
2. Allows the insertion, deletion and redirection of small DNA fragments
3. Free from the introduction of wide-range DNA damage, yielding higher safety
4. Can be applied in more cell line choices (including those that are hard to work with), and different editing circumstances
Prime Editing heralds a new era of genome editing, demonstrating significant potential in applications such as disease modeling, cell therapy, and gene function studies.
II. Prime Editing Mechanism & Rationale
How does Prime Editing work? It comprises 3 key components:
a. Prime Editing Guidance RNA (pegRNA) recognizes target nucleotide sequence, and encode a new DNA designed to substitute the target DNA sequence. Built upon an sgRNA, the pegRNA consists of the primer binding site (PBS) and the RT template (RTT). During gene editing, PBS allows for hybridisation between the nicked 3’ strand and the pegRNA, while RTT serves as a template for later DNA synthesis.
b. The H840A nickase is fused with a M-MLV-derived Reverse Transcriptase (RT), in which the H840A nickase is a derivative of Cas9 with a H840A mutation, which results in a loss of function of the HNH domain, leaving only the RuvC carrying out its cutting function. Having lost one of its cutting domain, the engineered enzyme can only form a single-strand “nick” on the DNA, hence the name nickase. M-MLV-RT, on the other hand, is capable of formulating DNA from ssRNA template.
c. Single Guidance RNA (sgRNA) operates so that the nickase can be navigated to the target DNA site.
Prime Editing Protocol:
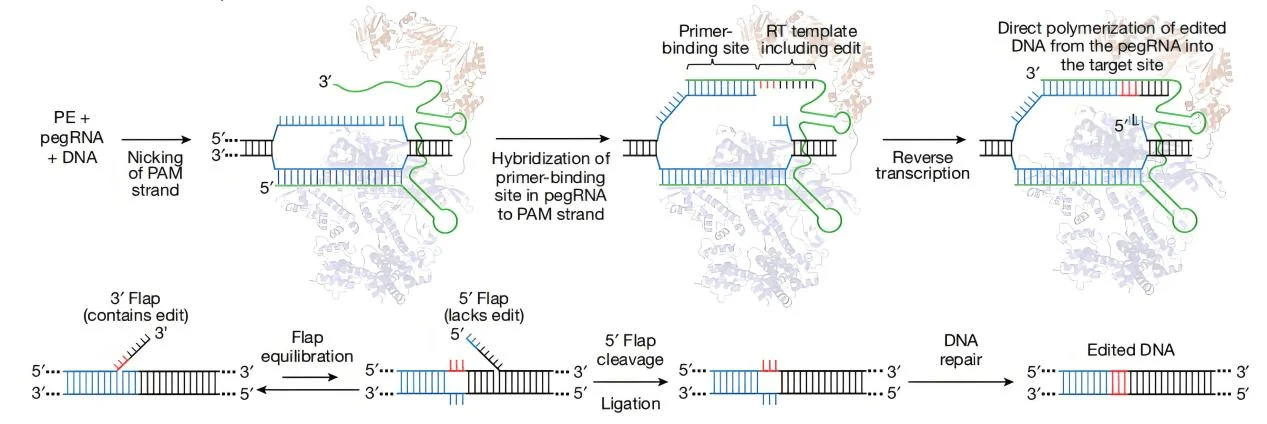
Figure 1. Fundamental principles of Prime Editing,(Anzalone et al.,2019)
During genome editing, the pegRNA and the protein complex are typically transfected into cells using a vector. The fusion protein cleaves the target DNA sequence, creating a single strand break and exposing the 3'-hydroxyl end. This end can initiate the reversed transcripting process of the RT template part in the pegRNA, forming a branched intermediate. This intermediate contains two DNA strands: a 3' strand with the newly synthesized (edited) sequence and a 5' strand with the unedited DNA sequence. Subsequently, the 5' strand is cleaved by a site-specific endo- or exonuclease, allowing the 3' strand to be ligated, forming a heterozygous double-stranded DNA, composed of an edited strand and an unedited strand. There are nucleotide mismatches at the editing site in the re - annealed double - stranded DNA will have nucleotide mismatches, which will trigger the cell’s own repairing mechanism. This may lead to two outcomes, with either the reincorporation the original nucleotides into the edited strand, resulting in an edit failure, or having the desired copying of the information from the edited strand onto the complementary strand, successfully stablising the edit.
Prime Editing (PE) is designed so as to be less reliant on DSBs, thereby reducing potential risks such as chromosomal abnormalities to certain extent. Thanks to its unique "search and replace" mechanism, Prime Editing technology can cover approximately 89% of known pathogenic mutations and support up to 12 types of base conversions and carry out precise editing of small fragments. It promotes the diversity and precision of gene modification, providing new possibilities for personalised medicine and cell model construction.
III. Prime Editing Evolution Timeline
| Versions | Time developed | Characteristics | Mechanisms of Action |
|---|---|---|---|
| PE1 | 2019 | PE1 can achieve basic editing but with low efficiency and non-specificity | pegRNA forms a complex with nCas9, locating the target DNA and binds. nCas9 produces a nick for the 3’ end of the cut to be joined with PBS, activating RT and synthesising DNA using the RTT. The edited sequence will compete with the original to form a double strand, with the outcompeted simple cleaved by cell mechanisms. |
| PE2 | 2019 | Reconstructed the M-MLV-RT, with a 1-5 increased editing efficiency over PE1 | 5 amino acid changes as compared to that in PE1, enabling stronger DNA-RNA binding with better stability and heat sustaining capability. Tighter binding with pegRNA accelerates reverse transcription and improves editing efficiency. |
| PE3 | 2019 | Introduces extra sgRNA with theoretically improved efficiency, but also risk of having deletions. | One more sgRNA on top of PE2, redirecting the Cas9 to cut DNA strands with complimentary editing sites. Expected with elevated efficacy but causes DSB in practice, triggering HDR, resulting in indel. Research showed a 1.5-4.2 multiplier comparing PE3 with PE2 efficiency. |
| PE3b | 2019 | PE3b redesigns the sgRNA to avoid the indel problem of PE3. | Revisited the sgRNA design by adding editing sequences.Through the induction of mismatch repairing mechanisms, PE3b accurately introduce edits while eliminating indel by inducing precise repair of the complimentary strand using the editing sequence. |
| Twin PE | 2021 | Twin prime editing can edit longer fragments with higher precision, yielding therapeutic potential | Twin prime editing utilises 2 pegRNAs, to form single strand break at different cutting sites to avoid DSB. The integration of site-specific recombinases also enables installation of "safe harbour" for the insertion, replacement or deletion of longer fragments |
| PE4、PE5 | 2020 - 2021 | Expressing the MLH1dn protein to reduce mismatching repair and indel genotypes | DNA mismatch repair (MMR) mechanism can reduce the efficiency of prime editing and increase indel by-products. Research has found that knocking down MMR-related genes (such as MSH2, MSH6, MLH1, and PMS2) can significantly improve the efficiency of prime editing. The PE4 and PE5 systems have been thus developed to co-expressing dominant-negative MLH1 (MLH1dn) to inhibit MMR, the editing efficiency of PE4 and PE5 systems in various cells is 7.7-times and 2-times higher than that of PE2 and PE3 respectively, and the edited/indel ratio is also improved. In addition, introducing silent mutations near the target editing site can evade MMR recognition and further improve the efficiency of prime editing. |
| PEmax | 2024 | PEmax shows a 2.5-fold increase in editing efficiency compared to PE2 in HeLa cells, and 1.2x more efficient in 293T editing. | PE2 upgraded in multiple aspects, optimised RT codon to have better transcription, improved cutting efficiency through engineering the spCas9 aa sequence. NLS peptide added on both ends of the enzyme for better affinity and a quicker editing process, ultimately increasing editing efficiency |
| PE7 | 2024.4 | Compared to PEmax, PE7 achieves over a 10-fold increase in editing efficiency in the U2OS cell line, PE7 increases the success rate of point mutations by 20% compared to PE5. 1.76-4.71x efficacy in difficult target sites, and 86% improvement in regular editing sites. | The Princeton team discovered that the small RNA-binding exonuclease protection factor La is the key. PE7 fuses La with the prime editing protein and uses an optimized pegRNA. La can bind to the polyuridine at the 3' end of the nascent RNA polymerase III transcript and interact with the 3' end of pegRNA to improve PE in both efficacy and variety, making PE7 a stronger version of PE. |
| uPEn | 2025 | uPEn achieves high editing efficiency and enables multiplex editing, holds great potential for livestock breeding | Integrates i53 module to optimise DNA repairing environment. uPEn coordinates pegRNA and enzymes to carry out precise induced repair and elevated efficiency in cell and zygote editing. Can target and act on multiple gene at the same time, such as the dual editing the the MSTN KO and KI of the Kozak sequence into the PPARG gene in sheep. |
Chart 1. Differences between different versions of Prime Editing
IV. Prime Editing Application Prospects
Prime Editing has a wide application spectra across multiple fields due to its feature of high efficacy, precision and low off-target rate.
1.Therapeutics
Therapy for single-gene disorders:
PKU is resulted by mutated PAH gene. A well-designed Prime Editing strategy, with pegRNA guiding the nCas9 to introduce the cut and RT to facilitate the synthesis of WT PAH sequence, was shown in murine model to revert phenylalanine metabolism back to normal, with 40% of the mice having recovered PAH activity and subsided Phe level.
Sickle cell Anemia is caused by a mutation in the HBB gene. Prime Editing technology was shown to have a 20-30% rate of successful edits in some PM attempts on patient-derived hematopoietic stem cells (HSCs) in an April 2023 issue of Nature Biomedical Engineering, providing new perspectives for therapeutic proposals in sickle cell disease.
2.Gene function researches
Studies in which BRCA1 was targeted by Prime Editing in breast cancer cell lines, showed a 70% cells carrying the anticipated edit, while also exhibiting significant difference from unedited cells in terms of proliferation migration in vitro. Such finding has empowered gene function researches of the tumour with a firm factual ground.
3.New Drug Development
In a lung cancer research focused on EGFR mutations, cell lines carrying a EGFR-sensitive mutation was constructed via Prime Editing technologies, yielding a new drug candidate with 80% and up targeted suppresion rate, way higher than that in traditional medications. Besides, Prime Editing can optimise the drug target site through precisely editing to modify its affinity or sensitivity to the drug. Thus, substances with higher potency and higher curing rate could be developed while new approaches and research topics also emerges.
4.Agricultural Breeding
Prime Editing was applied in rice breeding to produce plants with more favoured characteristics, such as lodging resistance and increased output. 30% of the GM plant was shown to have better lodging resistance and 10-15% showed a higher output, following SD1 gene editing by Prime Editing, showing potential for a novel breeding technique. With the help of an updated Prime Editing, uPEn, Professor Yongjie Wan and Professor Yanli Zhang’s team published their paper on Protein & Cell, in which they performed a double edit of the MSTN and PPARG genes in sheep, prospecting new sheep breeds with better growth and more delicious meat.
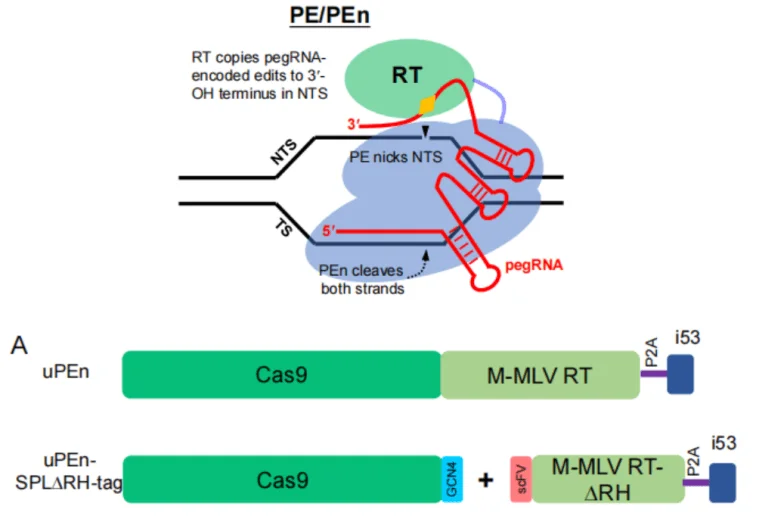
Figure 2. NJAU and NJU collaborate to develop an upgraded Prime Editing tool, uPEn
V. The Take-home Message
Against the backdrop where traditional gene editing technologies have laid a solid foundation for life sciences research and applications, Prime Editing (PE), a novel and precise editing tool that does not rely on DNA double-strand breaks or donor templates, displays broad application prospects. From PE1 to PE7 and the upcoming development of its derivative strategies, Prime Editing has been continuously optimized in terms of editing efficiency, precision, and operational flexibility, thereby expanding its application scope in multiple fields, including therapeutics, gene function research, new drug development, and agricultural breeding.
Looking ahead, combining the improvement of pegRNA design, the enhancement of RT performance, the deepening of understanding of DNA repairing mechanisms, and the innovation of delivering methodologies (nanocarriers, target-specific viral vector, etc.), Prime Editing is expected to further enrich the genome editing toolbox and provide strong support for the challenges in public health and sustainable development.
Want to know how Prime Editing technology can help your research? Come get in touch with our technical support now>>>
Reference:
[1] Anzalone, A. V., Randolph, P. B., Davis, J. R., et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature,2019,576(7785).
[2] Chen PJ, Hussmann JA, Yan J., et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021 Oct 28;184(22):5635-5652.e29.
[3] Yan J, Oyler-Castrillo P, Ravisankar P., et al. Improving prime editing with an endogenous small RNA-binding protein. Nature. 2024 Apr;628(8008):639-647.
[4] Mao W, Wang P, Zhou L., et al. An upgraded nuclease prime editor platform enables high-efficiency singled or multiplexed knock-in/knockout of genes in mouse and sheep zygotes. Protein Cell. 2025 Jan 20:pwaf006.
[5] Chen PJ, Liu DR. Prime editing for precise and highly versatile genome manipulation. Nat Rev Genet. 2023 Mar;24(3):161-177.
[6] Everette KA, Newby GA, Levine RM., et al. Ex vivo prime editing of patient haematopoietic stem cells rescues sickle-cell disease phenotypes after engraftment in mice. Nat Biomed Eng. 2023 May;7(5):616-628.


