From Scientific Question to Experimental Design: Leveraging CRISPR Screening to Elucidate the Regulatory Network of Pyrop


From Scientific Question to Experimental Design: Leveraging CRISPRS creening to Elucidate the Regulatory Network of Pyroptosis
Introduction: From Curiosity to Research Question — Why Do Cells Opt for Death?
Scientific inquiry often originates from a seemingly simple yet profound question. In the context of immunology and cell death research, this question can be posed as follows: why do some cells succumb rapidly while others survive when confronted with the same pathogenic stimulus? In recent years, as studies on programmed cell death have advanced, pyroptosis has emerged as a focus of intense interest. Pyroptosis is mechanistically and functionally distinct from classical apoptosis and passive necrosis, primarily due to its pro-inflammatory nature. At the molecular level, pyroptosis is characterized by the cleavage of Gasdermin proteins by inflammatory caspases. The resulting N-terminal fragments insert into the plasma membrane, forming pores that drive rapid cell swelling and lysis, accompanied by the release of potent pro-inflammatory cytokines, including IL-1β and IL-18.
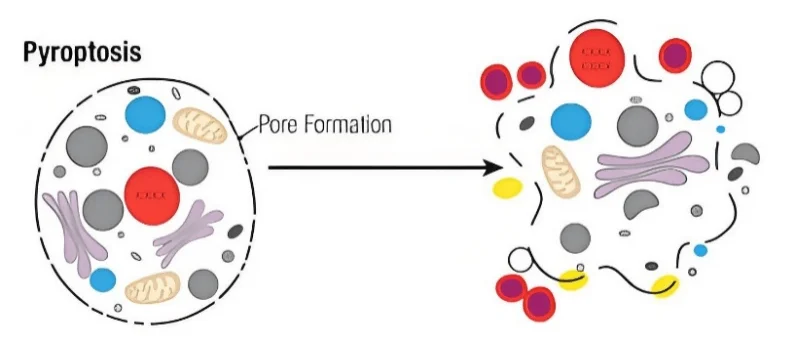
Figure 1. Pyroptosis in Cells
Pyroptosis plays a critical role in defending against intracellular pathogens, including bacteria, viruses, fungi, and protozoa, by eliciting a robust local inflammatory response that enhances immune defense. However, excessive activation of pyroptosis can lead to tissue damage and chronic inflammation, and it has been implicated in the pathogenesis of certain autoimmune and neurodegenerative diseases. A central question that remains largely unanswered is: how is pyroptosis regulated, and which genes determine whether a cell will engage the pyroptotic pathway upon encountering pathogens or stress?
Traditional molecular biology approaches, which typically focus on single genes, are often insufficient to address such system-level questions. To unravel the regulatory mechanisms underlying pyroptosis, higher-throughput and unbiased strategies are required. CRISPR screening represents a powerful approach in this regard, enabling the perturbation of tens of thousands of genes in a single experiment and the rapid identification of key regulators through phenotypic selection. In this study, we use the example of screening for genes regulating pyroptosis to illustrate how a scientific question can be translated into a systematic, CRISPR-based experimental design, and we outline a scalable methodological framework for such investigations.
From Question to Experiment: Identifying Regulators of Pyroptosis
Posing a scientific question is only the starting point of research; the key lies in translating the question into a tractable experimental strategy. Taking pyroptosis as an example, researchers must first define the core features of the process, namely, as an inflammatory form of programmed cell death, it is characterized by plasma membrane rupture, release of pro-inflammatory cytokines, and potent immune activation. Based on these characteristics, the research objective can be further refined to the task of “identifying factors that regulate the occurrence of pyroptosis.”
1.Deconstructing the Pyroptotic Pathway: From Upstream Signals to Downstream Execution
Achieving this goal requires breaking down the complex biological process into distinct modules. The upstream involves the recognition and assembly of inflammasomes (e.g., NLRP3, AIM2, NLRC4); the midstream comprises the activation of inflammatory caspases (Caspase-1/4/5/11); and the downstream entails Gasdermin cleavage and pore formation, accompanied by the maturation and release of IL-1β and IL-18. Researchers need to pinpoint which molecules within these modules have not yet been systematically explored.
2.Selecting a CRISPR Library: From Phenotype to Key Genes
CRISPR library screening provides a natural fit for addressing this scientific question. Pyroptosis exhibits a clear binary phenotype—cell death or survival—which is particularly suitable for survival-based screening strategies. Following the identification of candidate genes from the library screen, these hits are further validated through single-gene knockout, protein-level assays, and functional experiments, ultimately allowing reconstruction of the regulatory network.
Thus, the original scientific question is transformed into a clear experimental path:
- - Establish screening conditions via pyroptosis induction
- - Perform CRISPR library screening
- - Analyze sequencing results to identify candidate genes
- - Validate candidates individually and reconstruct the regulatory network
Unveiling Pyroptosis: The Molecular Story Behind Membrane Rupture
Rational experimental design requires a deep understanding of the biological underpinnings of pyroptosis itself. Pyroptosis is an inflammatory caspase-dependent form of programmed cell death, whose core mechanism revolves around the cleavage of Gasdermin proteins and the formation of membrane pores. This process ultimately leads to cell swelling, plasma membrane rupture, and the release of pro-inflammatory cytokines IL-1β and IL-18, thereby triggering local and systemic inflammatory responses. Pyroptosis primarily occurs through two distinct pathways:
- - Canonical Pathway:This pathway depends on the activation of Caspase-1. Pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) are recognized by intracellular inflammasomes such as NLRP3 or AIM2, which then recruit and activate Caspase-1. Activated Caspase-1 cleaves pro-IL-1β and pro-IL-18 into their mature, secreted forms and simultaneously cleaves the key executioner protein GSDMD. The N-terminal fragment of GSDMD (GSDMD-N) oligomerizes and inserts into the plasma membrane, forming pores that cause the release of cellular contents and ultimately cell lysis.
- - Non-canonical Pathway:This pathway operates independently of Caspase-1 and is primarily triggered by intracellular lipopolysaccharide (LPS), which directly activates Caspase-4/5/11. The activated caspases then cleave GSDMD directly, leading to pore formation and pyroptotic cell death in a manner similar to the canonical pathway.
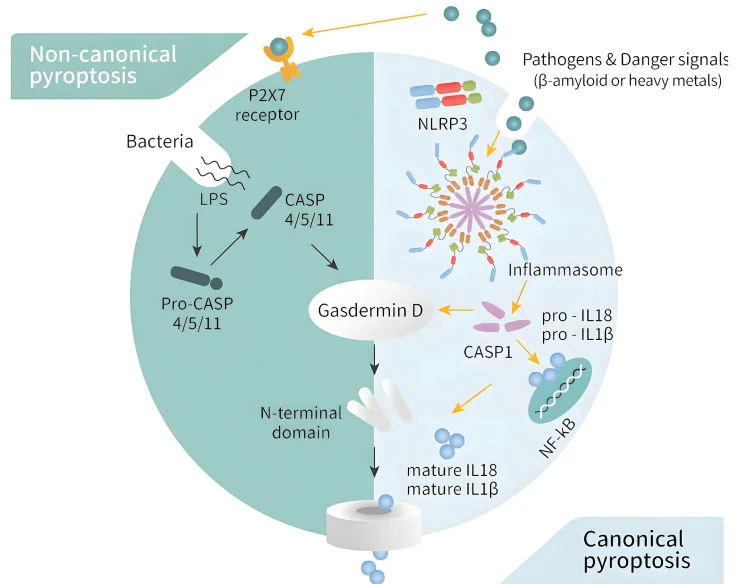
Figure 2. Two Core Pathways of Pyroptosis
From a molecular perspective, pyroptosis involves inflammasome activation, caspase activation, Gasdermin cleavage, and ultimately plasma membrane rupture accompanied by the release of pro-inflammatory cytokines. Inflammasomes such as NLRP3, AIM2, and NLRC4 detect pathogenic or cellular stress signals, recruiting and activating Caspase-1. In human cells, Caspase-4/5 mediates pyroptosis via the non-canonical pathway, whereas in murine cells, Caspase-11 performs an analogous function. Cleavage of GSDMD by these caspases releases the N-terminal fragment, which inserts into the plasma membrane to form pores approximately 10–20 nm in diameter. This pore formation disrupts ionic homeostasis, leads to cell swelling and lysis, and facilitates the passive release of inflammatory cytokines such as IL-1β and IL-18.
Visible Death: Membrane Rupture and Cytokine Release
Experimentally, pyroptosis exhibits clear and measurable hallmarks. Membrane rupture can be monitored via LDH release or PI/SYTOX staining, inflammasome assembly can be observed through ASC speck formation, and IL-1β/IL-18 secretion can be quantified by ELISA. These readouts not only provide temporal and quantitative insight into pyroptosis but also serve as reliable phenotypic indicators for subsequent gene screening and mechanistic validation.
Apoptosis vs. Pyroptosis
Compared with apoptosis, pyroptosis presents more pronounced phenotypes. Apoptotic cells shrink, maintain membrane integrity, undergo DNA fragmentation, and do not provoke an inflammatory response. In contrast, pyroptotic cells swell, rupture their membranes, and release large amounts of pro-inflammatory cytokines, triggering strong immune activation. These distinct differences make pyroptosis an ideal model for studying inflammatory cell death and its regulatory networks.
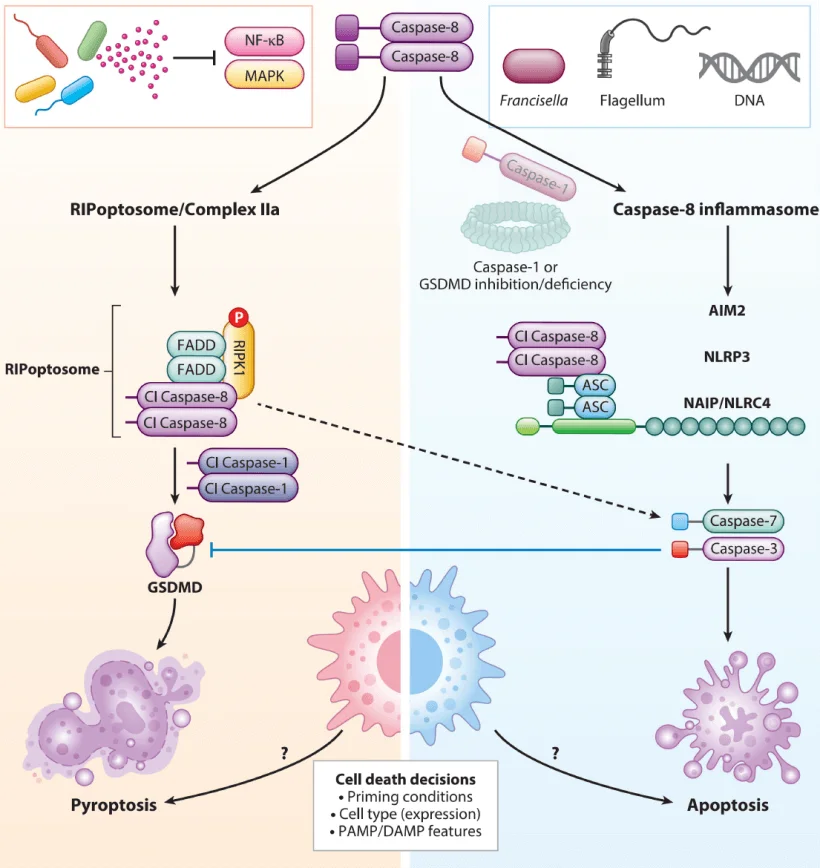
Figure 3. Key Differences Between Pyroptosis and Apoptosis
Adapted from Annu. Rev. Microbiol. 2023, 77:451–477.
How CRISPR Libraries Pinpoint Key Genes
CRISPR-Cas9 technology enables gene knockout by directing the Cas9 nuclease to specific genomic loci via guide RNAs (gRNAs). When expanded into library screening, this approach allows systematic evaluation of gene function at a genome-wide scale in a single experiment. A typical CRISPR library contains tens of thousands of sgRNAs, with each gene usually targeted by 3–6 independent sequences. In a screening experiment, the library is delivered into a population of cells stably expressing Cas9, typically via lentiviral transduction, and the cells are subsequently subjected to a defined selection pressure, such as pyroptosis induction.
Due to the binary nature of pyroptotic phenotypes, sgRNAs targeting genes whose loss confers survival are enriched in the surviving population. By extracting genomic DNA, amplifying sgRNA sequences, and performing high-throughput sequencing, changes in sgRNA abundance can be quantified, allowing the identification of candidate key regulators. The utility of CRISPR library screening for pyroptosis is particularly evident in three aspects: first, the binary phenotype makes survival-based screens highly efficient; second, the unbiased nature of the CRISPR library enables capture of both known and previously uncharacterized regulatory genes; and third, this strategy can be extended to study other biological processes, including apoptosis, ferroptosis, autophagy, and drug resistance.
From Design to Implementation: Experimental Strategies for Efficiently Screening Pyroptosis Regulatory Networks
When designing CRISPR library-based pyroptosis screens, the research objective should focus on constructing a comprehensive regulatory network, encompassing both positive and negative regulators. Regarding cellular models, THP-1 monocytes represent a classic system for studying pyroptosis. Common induction conditions include LPS + Nigericin to activate the NLRP3 inflammasome or electroporation of LPS to mimic the non-canonical pathway.
CRISPR Library selection typically employs genome-wide knockout libraries, such as Brunello or GeCKO v2, to ensure sufficient coverage and representation. In vitro experiments generally maintain a coverage of >500×, whereas flow cytometry-based screens commonly use ~300× coverage. Screening strategies can be categorized into two main types: first, survival-based screens exploiting differences in cell death and survival, suitable for rapid primary screening; second, flow cytometry-based refined screens, which use membrane permeability dyes, Caspase-1 activity probes, or ASC speck markers to distinguish different stages of pyroptotic cells, enabling identification of negative regulators and stage-specific modulators.
To ensure the reliability of screening results, experimental design must control multiplicity of infection (MOI) at low levels to ensure that each cell carries a single sgRNA, and non-infected cells should be eliminated through antibiotic selection. Additionally, maintaining an initial input sample as a reference is essential for subsequent statistical analyses.
Key Steps to Ensure Accurate and Reliable Screening
During experimental implementation, the first step is to establish a cell line stably expressing Cas9 to ensure high-efficiency genome editing. This is followed by library transduction and expansion, maintaining even sgRNA distribution and adequate coverage. Pyroptosis induction conditions must be pre-optimized to define an appropriate time window, typically 30–90 minutes post-stimulation, when characteristic membrane rupture and cytokine release can be observed.
During cell population collection, either surviving cells can be isolated directly, or early- and late-stage pyroptotic populations can be sorted by flow cytometry, depending on the experimental objectives. Subsequent steps include genomic DNA extraction, sgRNA amplification, sequencing, and data analysis. Commonly used statistical tools, such as MAGeCK, integrate enrichment signals from multiple sgRNAs to generate a list of candidate genes.
Quality Control Considerations: Quality control is critical throughout the screening process. Researchers should assess library distribution uniformity using the Gini coefficient and validate screening efficiency by confirming the significant enrichment of positive control genes, such as NLRP3, CASP1, and GSDMD. Additionally, correlation analysis across biological replicates is essential to ensure the reliability of the results.
From Candidates to Confirmation: Systematic Validation of Pyroptosis Regulators
The immediate output of a CRISPR library screen is a list of candidate genes, which must be validated through subsequent experiments. First, multiple sgRNAs can be designed for each candidate gene to generate single-gene knockout cell lines, with editing efficiency confirmed by protein-level assays such as Western blotting. Next, under the same pyroptosis-inducing conditions, functional assays—such as LDH release, IL-1β quantification, and inflammasome assembly assessment—can be performed to determine whether the phenotypes align with the screening results.
Further mechanistic studies can employ time-course experiments to elucidate the role of candidate genes at different stages of pyroptosis, or integrate pharmacological tools for pathway dissection—for example, using MCC950 to block NLRP3 or Disulfiram to inhibit GSDMD—thereby pinpointing the functional position of each candidate within the regulatory network. Cross-model validation is also essential, with experiments repeated in different cell lines or animal models to evaluate the generalizability of the findings.
A Generalizable Methodological Framework: Lessons from Pyroptosis
The study of pyroptosis provides a blueprint for establishing a generalizable methodological framework. First, clearly defining the scientific question and establishing well-characterized phenotypes is a prerequisite for successful screening experiments. Second, experimental design must carefully consider the choice of screening strategy and library coverage to ensure results possess both breadth and resolution. Third, rigorous quality control and appropriate control setups are central to the reliability of the data. Finally, CRISPR library screening results should be regarded as a starting point for hypothesis generation rather than a definitive conclusion, necessitating independent validation and cross-system verification to strengthen confidence in the findings.
This framework is not only applicable to pyroptosis research but can also be extended to investigations of apoptosis, ferroptosis, autophagy, and other forms of programmed cell death, as well as broader studies of drug resistance and disease mechanisms.
Conclusion
The essence of scientific research lies in starting from a question, selecting appropriate tools, and designing rational experiments to seek answers. Taking pyroptosis as an example, CRISPR library screening not only enables the identification of novel regulatory factors but also provides a generalizable methodological pathway: define the question, match the tool, control variables, validate hypotheses, and translate findings into applications. This approach is equally applicable to the study of other complex biological processes. From exploring fundamental mechanisms, dissecting disease pathology, to discovering new targets and developing therapeutics, CRISPR library screening is increasingly serving as a vital bridge connecting basic research and clinical application.
Ubigene | Comprehensive In Vitro and In Vivo CRISPR Screening
Ubigene offers end-to-end in vitro and in vivo CRISPRscreening services, supported by the newly developed iScreenAnlys™ CRISPR Library Analysis Platform, which streamlines the entire workflow from experimental design to data analysis. In vitro screens can be completed and delivered in as little as 6 weeks, while in vivo screens faithfully recapitulate the tumor microenvironment. The platform features zero-code operation and enables one-click generation of publication-ready figures, helping research teams efficiently and reliably identify high-value targets.
Contact us to learn more about technical support>>
Reference
1.Galluzzi, L., Vitale, I., and Aaronson, S. A., et al. (2018). Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ, 25(3), 486-548.
2.Frank, D., & Kagan, V. (2018). Pyroptosis: The Cell’s Self-Destruct Pathway. Springer.
3.Liu, Y., Li, S., and Chen, S., et al. (2024). S-palmitoylation of GSDMD by DHHC7 facilitates its membrane translocation and pyroptotic cell death. Nat Cell Biol, 26(2), 272-286.
4.Herrmann BI, Grayczyk JP, Brodsky IE. Collab or Cancel? Bacterial Influencers of Inflammasome Signaling. Annu Rev Microbiol. 2023 Sep 15;77:451-477.
5.Doudna, J. A., & Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science, 346(6213), 1258096.
6.Shalem, O., Sanjana, N. E., Hartenian, E., et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 343(6166), 84-87.



