CRISPR Screening Identifies Targets of Tumor Immune Evasion


CRISPR Screening Identifies Targets of Tumor Immune Evasion
Immunotherapy has transformed cancer treatment, yet its efficacy is often hampered by tumor-intrinsic mechanisms of immune escape. To uncover novel molecular determinants of this process, researchers employed the murine melanoma cell line B16 engineered to stably express ovalbumin (OVA) and performed a CRISPR-based metabolic gene library screen both in vitro and in vivo.
The screen identified Vdac2 as a critical mediator of tumor immune evasion. Mechanistically, Vdac2 functions as an immune signal–dependent negative regulator, suppressing interferon-γ (IFN-γ)–induced tumor cell killing and restraining inflammatory reprogramming within the tumor microenvironment. Loss of Vdac2 not only enhanced IFN-γ–mediated cytotoxicity but also activated the cGAS–STING pathway, amplifying antitumor immune responses. Importantly, targeting Vdac2 significantly improved the efficacy of immunotherapy, highlighting its potential as a therapeutic target.
Together, this study establishes Vdac2 as a central regulator of tumor immune escape and provides a strong rationale for leveraging Vdac2 inhibition to enhance current immunotherapeutic strategies.
Highlights
1.Establishment of a high-specificity immune response model
A tumor cell line stably overexpressing the OVA antigen was generated and combined with OVA-specific CD8⁺ T cells (OT-I cells) to construct a system enabling precise antigen recognition and cytotoxicity. This model markedly enhanced T cell–mediated antitumor activity.
2.Target identification across distinct immune backgrounds
By comparing screening outcomes in immunocompetent versus immunodeficient mice, the study effectively distinguished immune-dependent from immune-independent gene targets, enabling accurate identification of molecules associated with tumor immune evasion.
3.Integrated in vitro and in vivo screening strategy
The combination of cell-based in vitro screening with in vivo animal model screening increased both the accuracy and biological relevance of CRISPR library hits, providing a more robust approach for discovering bona fide immune regulatory targets.
CRISPR Screening Reveals Key Targets of Tumor Immune Evasion
To systematically uncover molecular mechanisms by which tumor cells evade immune surveillance, researchers applied a combined in vivo and in vitro negative-selection CRISPR library screening strategy using a B16 melanoma cell line stably expressing ovalbumin (B16-OVA).
1. In vivo screening strategy
- - Modeling:B16-OVA cells carrying the CRISPR library were implanted into both T cell–deficient mice (Rag⁻/⁻) and immunocompetent C57BL/6 mice.
- - Immune stimulation:On day 7 post-implantation, pre-activated OT-I CD8⁺ T cells (specific for the OVA antigen) were transferred into C57BL/6 mice to boost antitumor immune responses.
- - Data acquisition:By comparing sgRNA distributions in residual tumor cells from Rag⁻/⁻ versus C57BL/6 mice, immune-related candidate genes were identified.
2. In vitro CRISPR screening strategy
- - Experimental design:B16-OVA library cells were divided into two groups: the experimental group was co-cultured with activated OT-I T cells, while the control group was cultured alone.
- - Immune pressure:In the experimental group, OT-I T cells specifically recognized and killed B16-OVA cells, imposing a selective immune pressure.
- - Target identification:Differential sgRNA abundance between experimental and control groups revealed key genes that regulate tumor cell sensitivity to T cell–mediated cytotoxicity.

Figure 1. Schematic of CRISPR Screening Strategy
Further comparative analysis revealed a substantial overlap between the in vivo and in vitro screening outcomes. Notably, Vdac2, Pigu, Pigk, Fitm2, Pde4b, and Ext1 were consistently identified as significantly negatively enriched hits in both systems. This convergence indicates that loss of these genes enhances tumor cell susceptibility to T cell–mediated cytotoxicity, suggesting their potential roles as critical molecular targets in the regulation of tumor immune evasion.
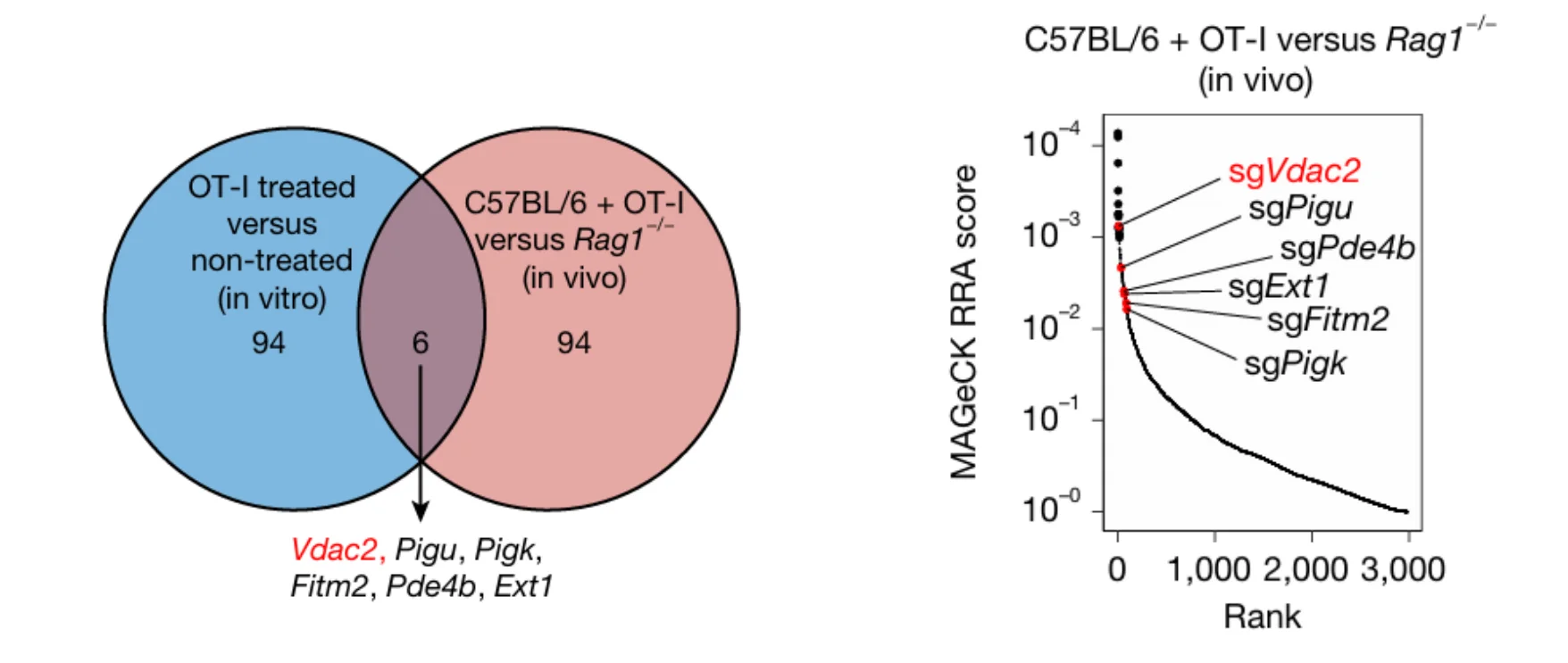
Figure 2. Identification of Immune Evasion Targets from In Vivo and In Vitro Screens
Validation of Vdac2 as a Regulator of Tumor Immune Evasion
From the initial CRISPR library screens, Vdac2 emerged as the most significantly negatively enriched gene, suggesting a pivotal role in tumor immune evasion. To validate this finding, researchers generated Vdac2 knockout B16-OVA cell lines and assessed their function through in vitro T cell co-culture assays and in vivo mouse transplantation models.
The results demonstrated that Vdac2 deletion markedly enhanced OT-I T cell–mediated cytotoxicity against B16-OVA tumor cells. Moreover, in immunocompetent mice, loss of Vdac2 significantly suppressed tumor growth. Collectively, these findings establish Vdac2 as a key regulator of tumor immune escape, with its absence sensitizing tumor cells to immune-mediated clearance.
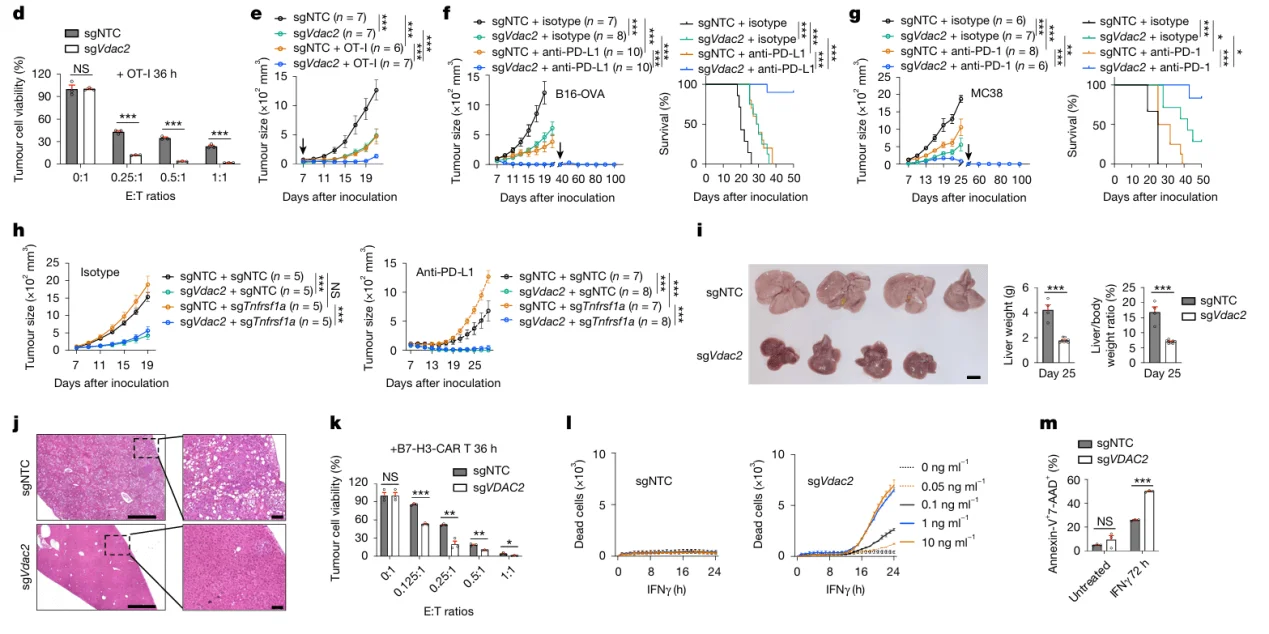
Figure 3. Validation of Vdac2 Knockout in Tumor Immune Evasion
Vdac2 Impairs IFN-γ–Induced STING Signaling
To elucidate the molecular mechanism by which Vdac2 promotes tumor immune evasion, transcriptomic profiling was performed on Vdac2-knockout versus control B16-OVA cells. The analysis revealed that loss of Vdac2 led to robust activation of multiple immune-related pathways, with marked upregulation of IFN-α and IFN-γ response genes. Furthermore, gene set enrichment analysis (GSEA) demonstrated a significant enhancement of cGAS–STING pathway activity in Vdac2-deficient cells.
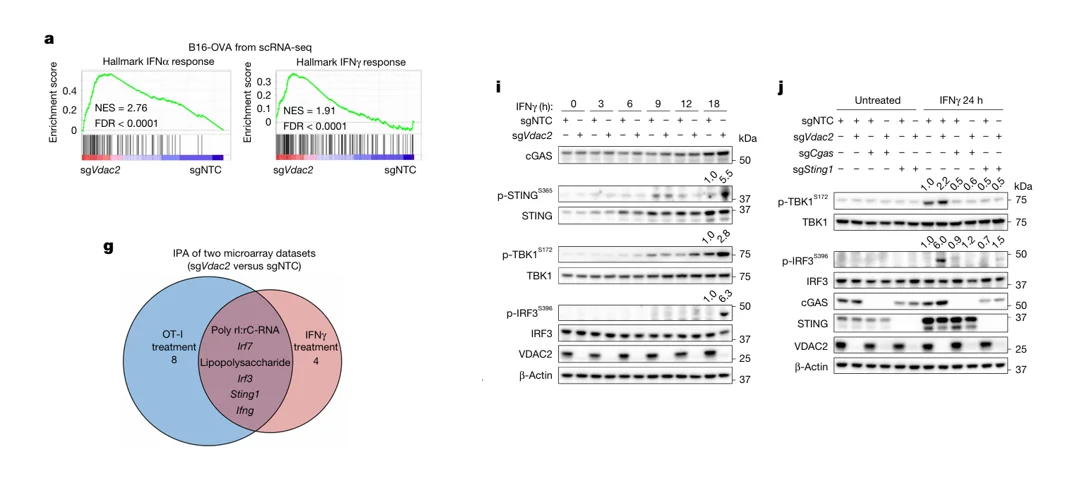
Figure 4. Vdac2 Impairs IFN-γ–Induced STING Signaling
CRISPR Screening Identifies IFN-γ–Dependent Sensitizer Genes
To further dissect the key mediators of IFN signaling–induced tumor cell clearance, researchers performed a genome-wide CRISPR knockout screen in Vdac2-deficient B16-OVA cells. During the screen, cells were stimulated with IFN-γ, and potential targets affecting IFN-γ–mediated cytotoxicity were identified by comparing sgRNA abundance between experimental and control groups.
The screen revealed significant enrichment of Casp9 and Bak1 (encoding the pro-apoptotic protein BAK). These findings suggest that loss of BAK impairs IFN-γ–induced tumor cell killing, further highlighting the critical role of the mitochondrial apoptosis pathway in IFN-γ–mediated immune clearance.
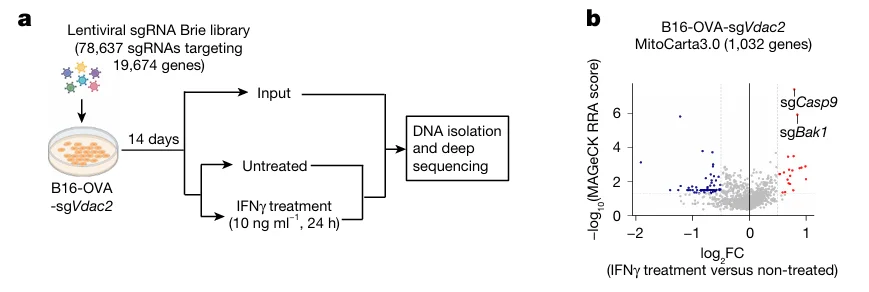
Figure 5. CRISPR Screening Identifies IFN-γ–Dependent Tumor Cell Death Regulators
Validation of BAK in IFN-γ–Mediated Tumor Cell Death
To validate the results of the CRISPR library screen, Bak1 was knocked out in B16-OVA cells, and its function was assessed in in vitro T cell co-culture assays as well as in vivo mouse tumor transplantation models. The results demonstrated that Bak1 deficiency markedly attenuated IFN-γ–induced tumor cell death, as evidenced by reduced apoptosis and sustained tumor growth in immunocompetent mice.
These findings further confirm that BAK is a critical mediator of IFN-γ–dependent tumor cell apoptosis, and its loss confers an immune evasion advantage to tumor cells.
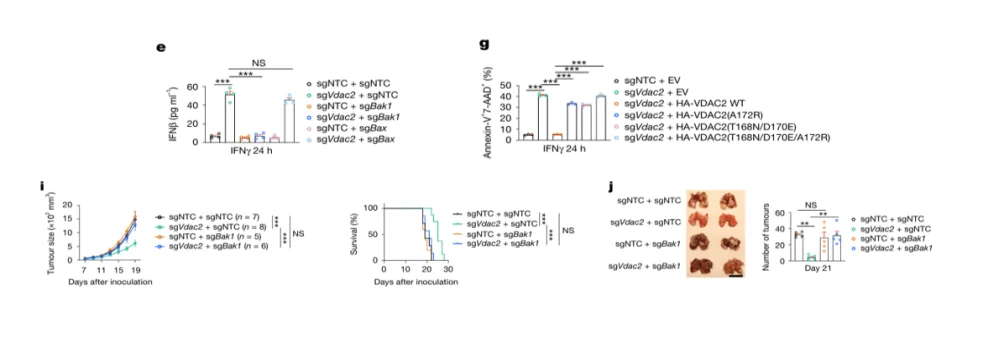
Figure 6. BAK Knockout Inhibits IFN-γ–Mediated Tumor Cell Death
New Semester, New Beginnings, New Discoveries! Ubigene proudly launches its dual in vitro + in vivo CRISPR screening service, now fully integrated with the iScreenAnlys™ platform—accelerating target discovery faster and smarter than ever!
- - CRISPR Library In Vitro Screening– Over 400 screening-ready cell pools, results in as fast as 6 weeks. Start screening immediately with no waiting.
- - CRISPR Library In Vivo Screening– Evaluate candidate genes in authentic physiological and tumor microenvironments, accurately reflecting tumor-immune interactions.
- - iScreenAnlys™ CRISPR Library Analysis Platform– No coding required. Automated QC, MAGeCK statistics, pathway enrichment, and publication-ready visualizations: target ranking heatmaps, differential gene volcano plots, and more. Ideal for manuscript drafts or report templates.
From experimental design to data analysis, Ubigene provides comprehensive support for universities, research institutes, and medical research centers. Turn in vitro and in vivo gene screening from a high-barrier challenge into a powerful research tool—accelerating experiments, enhancing reliability, and maximizing the value of target discovery.
Get in touch for expert technical support →
Reference
Yuan S, Sun R, Shi H, Chapman NM, Hu H, Guy C, Rankin S, Kc A, Palacios G, Meng X, Sun X, Zhou P, Yang X, Gottschalk S, Chi H. VDAC2 loss elicits tumour destruction and inflammation for cancer therapy. Nature. 2025 Apr;640(8060):1062-1071.



