CRISPR Screening Service
CRISPR libraries are high-throughput gene screening platforms based on the CRISPR/Cas9 system. Through functional screening, enrichment, and deep sequencing analysis, this approach enables the identification of phse, high knockout efficiency, and minimal off-target effects, CRISPR library screening has become a preferred platform for large-scale functional genomic studies.
Exploring the CRISPR/Cas9 Library: Precision Targeting to Uncover Essential Genes>>Based on its proprietary CRISPR-iScreen™ technology, Ubigene provides comprehensive, one-stop CRISPR screening services, including CRISPR-KO, CRISPRa, and CRISPRi customized libraries. The platform supports the entire experimental workflow — from high-throughput sgRNA library construction, viral packaging, and cell transduction, to drug selection, high-throughput sequencing, and bioinformatic data analysis.In addition to in vitro studies, Ubigene also offers in vivo CRISPR screening services to enable more physiologically relevant target discovery. Multiple delivery options are available to meet diverse research needs.
Ubigene CRISPR-iScreen™ Technique
CRISPR-iScreen™ is antechnology independently developed by Ubigene, designed to enable efficient and precise CRISPR screening. This technology provides researchers with a powerful and reliable tool for gene function studies and drug target discovery.
Proprietary High-Efficiency Competent Cells
Ubigene's specially developed competent cells exhibit superior DNA uptake capacity, enabling high transformation efficiency while minimizing mutation risk. This technology ensures robust plasmid library amplification with > 99% coverage and < 10% deviation in uniformity, providing a reliable foundation for high-quality CRISPR screening.
Fully Optimized Cell Pool Preparation Process
Ubigene's fully optimized Cell Pool preparation process allows scalable and standardized generation of CRISPR library Cell Pools, characterized by low inter-batch variability and excellent reproducibility. The Cell Pool libraries exhibit up to 99% coverage, ensuring consistency and reliability for downstream functional screening.
Diversified CRISPR Screening Platforms
Ubigene provides end-to-end CRISPR screening services for both in vitro and in vivo applications. Our platform enables flexible configuration of diverse experimental conditions, including compound treatment, passaging, viral infection, and flow cytometry–based selection. We also offer multiple enrichment strategies, empowering researchers to DIY their own functional screening systems. This highly adaptable platform is built to meet the evolving needs of cutting-edge life science research.
iScreenAnlys™ CRISPR Library Analysis System
This is an interactive, in-house developed analysis platform featuring an intuitive, user-friendly interface that requires no coding experience. The platform supports multiple statistical methods and a wide range of customizable data visualizations, enabling personalized analysis workflows. Users can generate publication-ready figures and reports, accelerating data interpretation and scientific discovery.
CRISPR Screening Services Overview
CRISPR Library Plasmid
Ubigene's CRISPR library plasmid services encompass gRNA design, primer array synthesis, vector assembly, plasmid preparation, and NGS-based validation, ensuring >99% coverage and <10% uniformity deviation. A portfolio of 40+ off-the-shelf library plasmids is also available, enabling flexible experimental design and supporting high-throughput CRISPR screening.
Virus & Cell Pool
Ubigene employs a third-generation, high-safety viral packaging system to produce viruses with titers of ≥1×10⁸ TU/ml. Low multiplicity-of-infection (MOI) transduction enables the construction of high-quality pooled cell libraries, supporting direct drug screening. Over 400 off-the-shelf library Cell Pools are also available, providing flexibility and efficiency for large-scale functional screening experiments.
In vitro Screening
By integrating various selective pressures and phenotypic enrichment methods, Ubigene develops tailored functional screening pipelines. The multifaceted phenotypic screening platform enables precise adaptation to diverse experimental requirements, supporting high-throughput and efficient discovery of critical genetic targets.
In Vivo Screening
In vivo CRISPR library screening allows the faithful replication of physiological microenvironments and dynamic biological processes, facilitating the accurate determination of gene function in vivo. Ubigene has successfully executed multiple in vivo screening studies, delivering reliable technical support for functional genomics and therapeutic target discovery.
NGS Sequencing
DNA is extracted from Baseline, negative control (NC), and experimental samples for next-generation sequencing (NGS) and gRNA differential analysis. Ubigene's platform facilitates the accurate identification of candidate targets, providing robust support for functional genomics and drug discovery studies.
CRISPR Screening Workflow
Prepartion Stage

gRNA design

Chip synthesis of Oligo Pool

Plasmid construction

Lentivirus packaging
Screening Stage

Cell Pool Construction (Cell infection by lentiviral library)

Cell screening (Positive or negative screening byapplying screening pressure)
Analysis Stage

PCR amplification

Sequencing

Analyze the data and find out candidates
Prepartion Stage
gRNA design
Chip synthesis of Oligo Pool
Plasmid construction
Lentivirus packaging
Screening Stage
Cell Pool Construction (Cell infection by lentiviral library)
Cell screening (Positive or negative screening byapplying screening pressure)
Analysis Stage
PCR amplification
Sequencing
Analyze the data and find out candidates



Case Studies
Screening functional genes of tumor
Negative screening of gene pairs with synthetic lethality
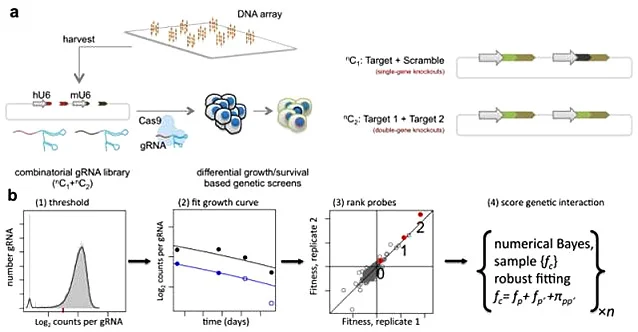
Synthetic lethality refers to the phenomenon that when two non-lethal mutant genes occur alone, they will not cause cell death, but when they occur simultaneously, they will cause cell death, which is one of the new research directions in the field of antitumor drugs. Because tumor cells often carry many point mutations, how to specifically kill tumor cells with high mutation rate without affecting normal cells is a major pursuit of antitumor drug research and development. Starting from the idea of synthetic lethality, Shen et al [2] designed a dual-gRNA library to screen the synthetic lethal interaction network. Different from the general sgRNA library, each vector in the dual-gRNA library contains two gRNAs, one targeting common mutated tumor suppressor genes in tumors and the other targeting genes that can be perturbed by anticancer drugs. They used this system to screen 73 genes in three experimental cancer cell lines (human cervical cancer HeLa, lung cancer A549 and embryonic renal cell carcinoma 293T), with a total of about 150000 gene combinations. By detecting gRNA abundance changes at different time points, they further analyzed and screened 120 synthetic lethal interactions, providing new targets for the development of new cancer drugs.
View Picture
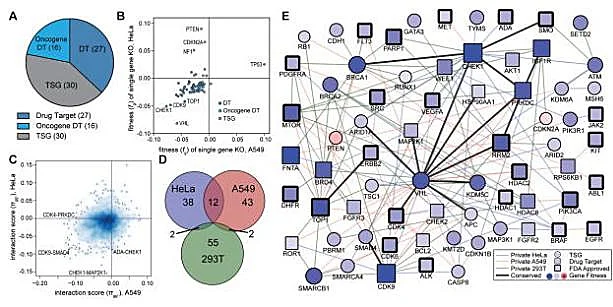
View Picture
Viral infection related study
Screening of HIV therapeutic targets
AIDS caused by HIV infection is a serious threat to human life and health. It is of great significance to clarify the molecular mechanism of HIV breaking through the host cell defense system and develop new targets for HIV treatment. Park et al. [3] infected cas9 virus on CCRF-CEM cells stably expressing CCR5 hygR and HIV-1 LTR-GFP, and screened a clone (GXRCas9 cell) that highly expressed CCR5 before HIV infection and low expressed EGFP but high expressed EGFP after infection as a tool cell for screening HIV infection targets. Specifically, GXRCas9 cells were infected with a lentiviral library containing 187536 sgRNAs (targeting 18543 genes), and these T cells lacking different receptors were infected with the HIV virus strain JR-CSF. Then, GFP negative and positive T cells were sorted out by flow cytometry, and the GFP negative cell population and the uninfected HIV virus cell population were sequenced to analyze the difference in sgRNA abundance between the two groups of cells. Finally, five genes with the largest change in sgRNA abundance were screened out. Among them, CD4 and CCR5 are the receptors of HIV-infected T cells. TPST2 and SLC35B2 modify CCR5 to facilitate the binding of HIV, while the gene encoding leukocyte adhesion factor ALCAM is related to the transmission of HIV between cells. The five genes screened do not affect the survival of T cells after being knocked out, but can make T cells resist HIV infection. Therefore, these five genes can be used as potential targets for HIV treatment, providing new ideas and ways for the prevention and treatment of AIDS.
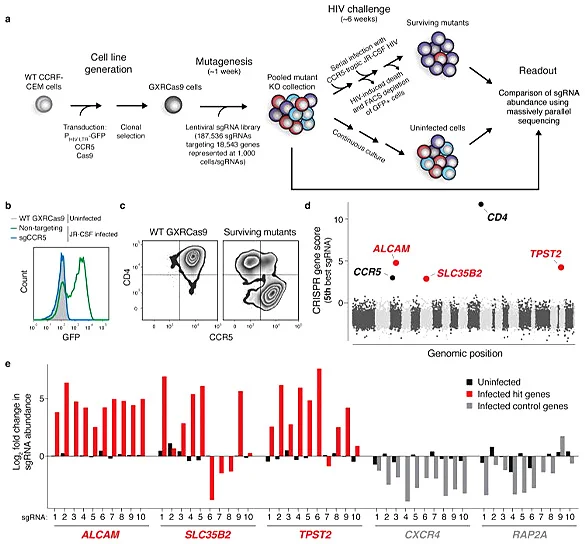
View Picture
Antibody target screening
Recognition of monoclonal antibody specific target antigens and their epitopes
It is quite simple to use peptides or purified proteins to verify antibodies in immune experiments, but it is relatively difficult to use whole cells or other complex antigens to verify antibodies. If no antibody reactivity is detected in Western blotting and immunoprecipitation, a variety of gene manipulation level techniques need to be applied to determine the antigen specificity of mAbs. BF4 is an antibody that can bind to the viral biofilm on the surface of uninfected lymphocytes, neutrophils and HTLV-1 infected cells. Zotova et al [4] used MT2 cells (human T cell lymphotropic virus type I HTLV-1 chronically infecting T cells) as immunogen to trigger mouse immunity and obtained a HTLV-1 biofilm specific monoclonal antibody BF4. Based on the idea of screening BF4 antigen knockout cells by transducing CRISPR knockout library into BF4 positive cells, they transduced GeCKO library to CEM T and Raji/CD4 B cells to sort out cells that did not bind to BF4. After two rounds of repeated sorting, the proportion of negative cells reached more than 99%. Researchers sequenced these cells and found that about 80% of sgRNAs targeted CD82. BF4 was confirmed to be a specific antibody against CD82.
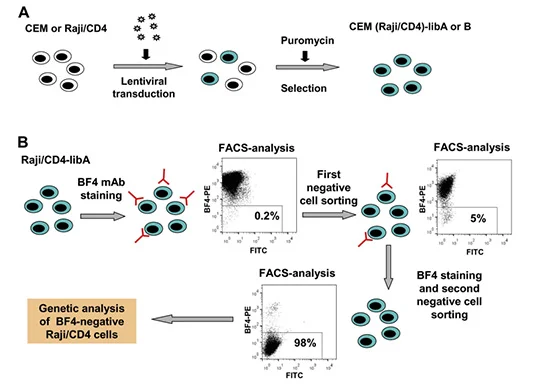
View Picture
Signal pathway
Studying the mechanism of pyroptosis
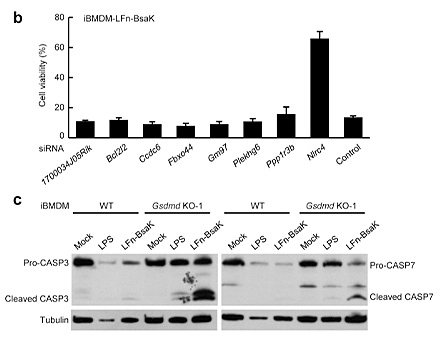
Pyroptosis is an immune defense response initiated by the body after sensing the infection of pathogenic microorganisms. Inflammation-activated caspase-1 and caspase-4, Caspase-5 and caspase-11, which recognize bacterial lipopolysaccharide, can cause pyroptosis, but the mechanism remains unknown. Shi et al [5] first established a lipopolysaccharide (LPS) electroporation method that can induce pyroptosis in more than 90% of cells, and then transduced the CRISPR knockout library into Tlr4-/- iBMDMs that can normally respond to lipopolysaccharide stimulation, and sequenced the cells that survived pyroptosis induced by lipopolysaccharide. The analysis results showed that four of the five sgRNAs targeting gasdermin D (GSDMD) gene had the top 30 copy numbers, and two of them were in the top 10 positions. The subsequent results also further proved that the N-terminus of GSDMD could induce pyroptosis. In summary, researchers used CRISPR library to conduct genome-wide genetic screening, successfully screened the gene GSDMD that can inhibit pyroptosis after knockout, and clarified the molecular mechanism of GSDMD as an inflammatory caspase substrate protein that can trigger pyroptosis after being cleaved.
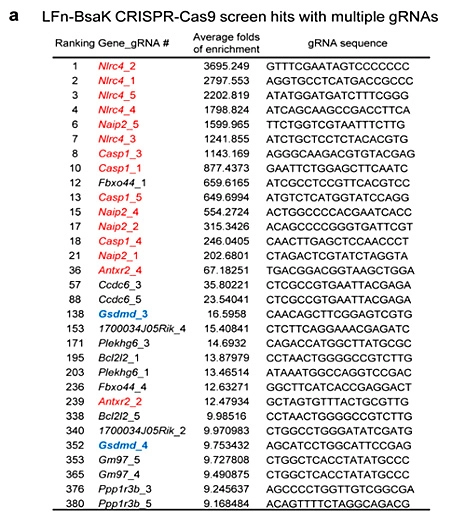
View Picture
View Picture
FAQs
1. Why is CRISPR screening important for gene function studies?
Traditional gene function studies mainly rely on forward genetics and reverse genetics. Forward genetics, often based on high-throughput multi-omics analyses, is expensive and makes it difficult to establish a direct link between gene targets and phenotypes. Reverse genetics involves direct gene perturbation at the cellular or organismal level, enabling clear causal relationships between genes and phenotypes. However, it can be costly, labor-intensive, and heavily dependent on prior knowledge. CRISPR library screening combines the high-throughput capacity of forward genetics with the causal clarity of reverse genetics, allowing researchers to unbiasedly identify phenotype-associated functional targets from large gene sets. This makes it a powerful and efficient tool for large-scale functional genomics research.
2. What are the key advantages of CRISPR library screening?
- Establishes a direct causal relationship between genes and phenotypes
- Enables unbiased, systematic functional exploration across a large number of genes
- Flexible and highly customizable screening systems
- Scalable with broad applicability of screening data
- Cost-effective and efficient for large-scale functional studies
- Features high specificity and low off-target effects
3. What research areas commonly use CRISPR library screening?
CRISPR screening is widely applied in oncogenesis, drug resistance, and viral infection studies to identify key genetic targets. It is also used to explore unknown gene functions in areas such as signaling pathways, immunology, and metabolism. In addition, CRISPR library screening can be adapted and optimized for more complex research systems, including stem cell models and aging studies, expanding its potential across a broad range of biological and disease research fields.
4. What is the CRISPR screening in T cells?
CRISPR screening in T cells uses gRNA libraries to target genes. T cells with altered genes are exposed to selection pressures like immune stimuli. This helps identify genes vital for T - cell functions such as activation and antigen recognition, advancing understanding of T - cell biology and immunotherapy.
5. What is the difference between RNA sequencing and CRISPR screen?
In summary, RNA-seq measures gene expression levels, while CRISPR screens identify functional roles of specific genes through genetic perturbation.
Learn more about their differences: Target Screening: CRISPR Library vs Traditional Omics
6. What is the result of the CRISPR screening?
CRISPR screening results in identifying genes linked to a specific phenotype. gRNA libraries target genes for disruption or activation. After selection based on the phenotype, gRNAs in surviving/altered cells are sequenced. In knockout screens, disrupted genes affecting survival are found; in activation screens, activated genes causing a phenotype are identified. This reveals gene functions, pathways, and potential therapeutic targets.
7. How long does it take to do a CRISPR screen?
The duration of a CRISPR screen can vary significantly depending on the type of screen and specific experimental conditions. Here are general timelines for positive and negative CRISPR screens:
- Positive CRISPR Screens• Typical Duration: 1-2 weeks.
- Negative CRISPR Screens• Typical Duration: 3-4 weeks.
Key Considerations
- Library Complexity: Ensure the initial cell population is large enough (e.g., 500x library complexity) to maintain sgRNA representation.
- Antibiotic Selection: Start antibiotic selection 72 hours after transduction to stabilize transduced cells.
- Genetic Drift: Avoid prolonged culture periods to prevent genetic drift, which can obscure results.
The exact duration may vary based on cell type, growth rates, and specific experimental conditions.
8. How much does the CRISPR library cost?