CRISPR gene-editing HeLa cells--front fighter of medical breakthroughs


CRISPR mediated HeLa cells--front fighter of medical breakthroughs
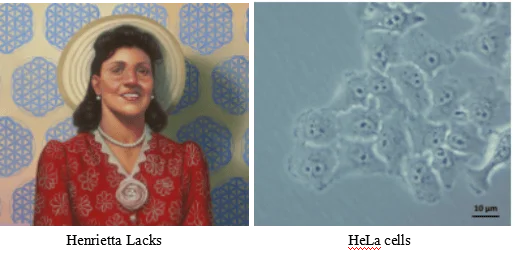
What are Hela Cells
In 1952, Henrietta Lack (HeLa) cells became the first human cell line that could grow and divide endlessly in a laboratory, leading scientists to label these cells "immortal". Researchers originally took HeLa cells from an aggressive cervical cancer tumor. These cells continue reproducing with a constant supply of nutrients, they produced a new generation of cells in less than 24 hours. Hela cells, like many tumors, have error-filled genomes, with one or more copies of many chromosomes: a normal cell has 46 chromosomes, where HeLa cells contain 76 to 80 total chromosomes, some of which are heavily mutated. These cells also expressed an over reactive telomerase that rebuilds telomeres after each division, preventing cellular aging and cellular senescence, and allowing perpetual divisions of HeLa cells. HeLa cell line has contributed to many medical breakthroughs, from research on the effects of zero gravity in outer space and the development of the polio vaccine to the study of leukemia, the AIDS virus, and cancer worldwide. Although many other cell lines are in use today, HeLa cells have supported advances in most fields of medical research in the decades since HeLa cells were isolated. HeLa cells have played in some of the major advances in fields such as cancer biology, infectious disease, fundamental microbiology, and many others. Research involving HeLa cells has been described in more than 110,00 scientific publications including three who have been awarded Novel Prize: in 2008 for the discovery that the human papillomavirus (HPV) is a causative agent of cervical cancer, in 2009 for the discovery of the telomerase enzyme, which protects the telomeres of the chromosome, thus preventing the degradation of the chromosome and in 2014 for advances in live viewing of cellular growth. This staggering number makes it clear just how important these cells have been to research over the past six decades.
Buy Hela cells for your research>>>
Why are Hela Cells Important?
The versatility and power of HeLa cells have made them an essential laboratory tool that still continues to offer new clues about the basis of human health and disease. HeLa cell one of the popular cellular model as well as tool for life scientists, who willing to study the mechanism of action of diseases or therapeutically active drug molecules as well as to decipher cell signaling events such as DNA damage repair. Researchers chooses HeLa cell as a tool for multidisciplinary studies due to its immortal cell characteristics, available multi-omics database (genomic, proteomic, and transcriptomic), and easy of to access these databases to design and modify research projects. One of the recent examples of using HeLa cells to study the infectivity of the virus SARS-CoV-2 in humans. Scientists studying COVID-19 using HeLa cells to determine the receptor the virus uses to enter human cells. The coronavirus study got an unprecedented pace, based on the studies rely on HeLa cells.
Application - What are Hela Cells Used for
HeLa cells have evolved into a useful model to study a huge variety of topics. HeLa cells derived from HPV-transformed cervical adenocarcinoma (cancer) cells. They have been used for studies including but by no means limited to those in cancer research. As a tumor model, they have been widely used in studies:
1. Tumor cell migration and invasion
2. Drug development
3. Cell death pathways
CRISPR-U™ Gene-editing in HeLa Cells
CRISPR-Cas systems have formed a revolutionary genome-editing tool, giving a great impetus to the development of life science and our understanding of life. The CRISPR system used for precise genome editing and can generate gene knockout or knock-in genome manipulations through substitution of a target genetic sequence with a desired donor sequence. HeLa cell is an aggressive cervical cancer cell line. Gene editing in HeLa cell line possible to generate single or multiple gene knockouts, correct mutations, or insert reporter transgenes. These genomes edited HeLa cell lines will allow investigators to study cancer research, cancer therapeutics, cell death research, functional genomics, signaling pathways, drug discovery, drug response, and cell therapy. Ubigene developed CRISPR-U™ for gene manipulation of HeLa cell line. Thereby, possible to achieve genome editing in HeLa cells with the utilization of the CRISPR/Cas9 system. Ubigene can customize the gene-editing in eukaryotic HeLa cells as well as can generate various genes modification in animal models.
Read more about Hela cells culture and gene-editing tips>>>
Download the Hela cell culture protocol for free>>>
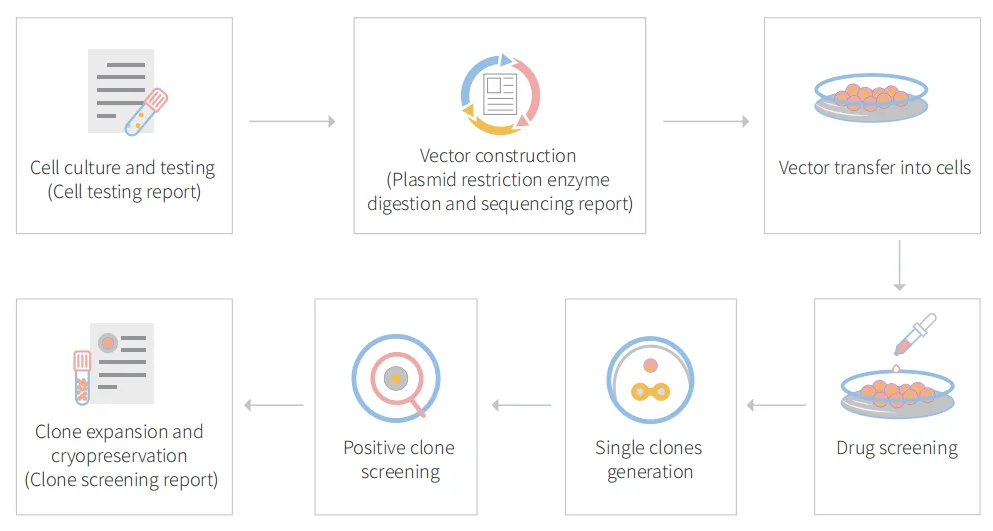
Figure: CRISPR-U™ customized workflow for engineered HeLa model cells
Ubigene developed CRISPR-U™ which optimizes eukaryotic cells and animal gene-editing vectors and process. The efficiency and accuracy are 10x higher than traditional methods. Contact us immediately to know about your research related services!
Case Study 1: Knockout
CRISPR/Cas9 mediated TERT disruption can suppress tumor cell survival
Telomeres are the unique structures in eukaryotic cells that maintain chromosome integrity. Mammalian telomere lengths are primarily regulated by telomerase, a ribonucleoprotein consisting of reverse transcriptase (TERT) and an RNA subunit (TERC). TERC is constitutively expressed in all cells, whereas TERT expression is temporally and spatially regulated in most adult somatic cells. In the somatic cell, TERT is inactivated and their telomerase activity is undetectable. Most tumor cells activate TERT as a mechanism for preventing progressive telomere erosion to achieve proliferative immortality. Therefore, inactivating TERT is consider to be a promising means of cancer therapy. The researcher applied CRISPR/Cas9 mediated gene-editing system to target the TERT gene in cancer cells. For this purpose, they selected three different cancer cell lines a cervical cancer cell line (HeLa), a pancreatic cancer cell line (PANC1), and a breast cancer cell line (SUM159). They designed three gRNAs (sg1, sg2, and sg3) targeting the human TERT gene exon 2 (E2), exon4 (E4) and exon 6 (E6), respectively. They further design another two gRNAs (sg4 and sg5) targeting the flanking introns of TERT exon 4 (E4) to generate KO TERT cells. In the haploinsufficiency study, the mutated TERT cells shown lower telomerase activity and shorter telomeres. The cell proliferation measurement, cell culture density, and stronger β-gal staining signals provide evidence that mutant TERT cells suffering severe cellular senescent compare to WTPE Hela cells. Annexin V and PI staining provide evidence about cellular apoptosis and, the TERT mutant shown higher apoptosis than WTPE Hela cells. The TERT haploinsufficiency in cancer cells leads to retarded growth and enhanced cell death In Vitro.

Fig1: Retarded growth and increased cell death in Tert+/− cancer cells. (A) Population doubling time of WT and TERT+/− Hela cells. (B) Light microscopy images of WT and TERT+/− Hela cells at Passage 2 (P2), P5 and P7. (C) β-gal staining of WT and TERT+/− Hela cells. Arrows: example of severely senescent cells. (D) Cell death rates of WT and TERT+/− Hela cells determined by LDH assay. (E) Quantification of flow cytometry analysis of annexin-V and propidium iodide (PI) staining of apoptotic cells in WT and TERT+/− Hela cells. ** p < 0.01.
They further studied on TERT haploinsufficiency has any effects on tumor cell survival in vivo using a tumor xenotransplant nude mice model. Six weeks post-inoculation examined the sizes of the xenografts. As expected, the WTPE Hela cells grew into tumor mass (diameters ~2 cm) in animals; where the TERT mutant Hela cells failed to form any tumor xenotransplant in animals. This result support that Cas9-mediated TERT haploinsufficiency can effectively suppress tumor cell growth in vivo.
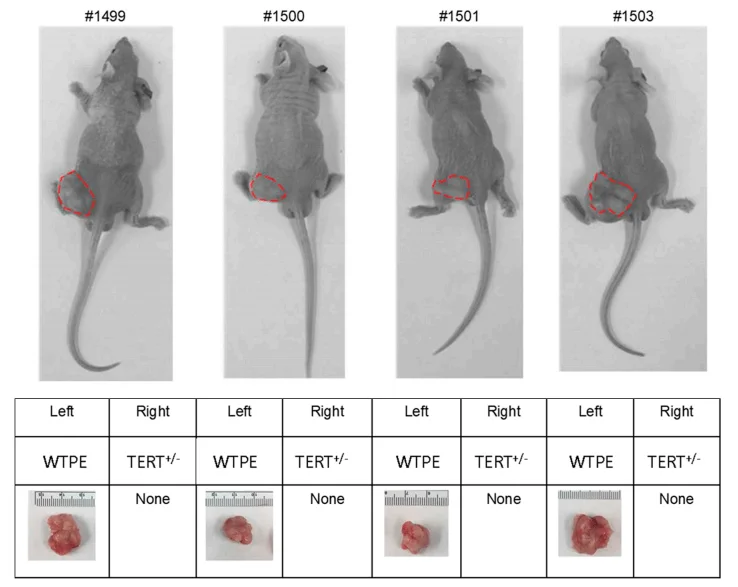
Fig 2. Xenotransplant of WT and TERT+/− Hela cells in nude mice
Case Study 2: Point Mutation
CRISPR/Cas9 introduces pathogenic MSH2 point mutations cause differential genomic de-stabilization in human cells
Repeat de-stabilization is variously associated with human disease especially in neoplastic diseases, microsatellite instability (MSI) has been regarded as simply reflecting DNA mismatch repair (MMR) deficiency. The MSI+ phenotype is not uniform in human neoplasms. Based on the frequency of microsatellite changes MSI are classified into MSI-H (high) and -L(low). Additionally, MSI also classified their distinct qualitative modes, i.e. Type A and Type B. An earlier study reported that tumors occurring in MMR gene knockout mice exhibited not drastic microsatellite changes typical in MSI-H tumors (i.e. Type B mode) but minor and more subtle alterations (i.e. Type A mode). Lynch syndrome (LS) known as hereditary non-polyposis colorectal cancer (HNPCC), is the most common cause of hereditary colorectal (colon) cancer. Mutation of these genes MLHL, MSH2, MSH6, PMS2, and EPCAM responsible for LS. In the present study, MSH2 mutations reported in Lynch syndrome (LS) were introduced into HeLa cells by using the CRISPR/Cas9 system. The researcher used genome editing vector, pSpCas9(BB)-2A-Puro (pX459), gRNA (designed by CRISPR direct and CRISPR Design Tool) and subcloned into pX459 using the BbsI site. The gRNA-encoding pX459 plasmid and three different single-stranded ssODNs were co-transfected into cells using Lipofectamine 2000. The established mutant clones clearly exhibited MMR-defective phenotypes with alkylating agent-tolerance and elevated mutation frequencies. However, microsatellites were not markedly destabilized as in MSI-H tumors occurring in LS patients and all the observed alterations were uniformly Type A, which confirms the results in mice. These findings suggest added complexities to the molecular mechanisms underlying repeat destabilization in the human genome.
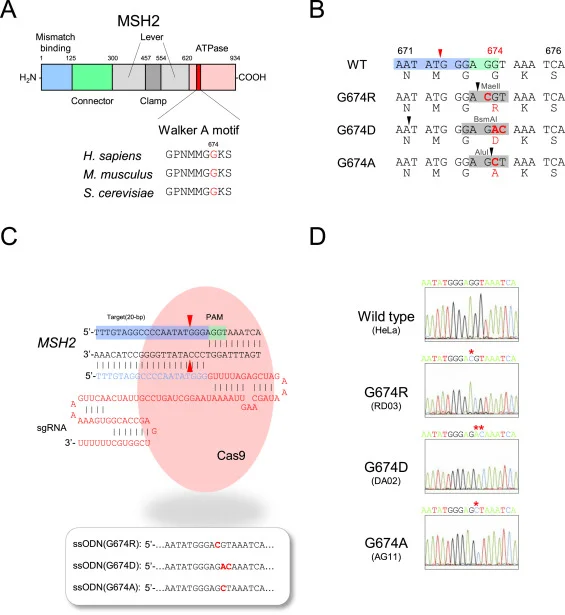
Fig. CRISPR/Cas9 system introducing MSH2 G674 mutations into HeLa cells. (A) Functional domains, including Walker A motif, in human MSH2 protein, (B) Local DNA and amino acid sequences in Walker A motif in each G674-mutant version, (C) The structure of the guide RNA and ssODNs used for study. (D)Sanger method confirm each of the established HeLa clones.
In this study, MSH2-mutant HeLa clones have been successfully obtained, and the established clones clearly exhibited MMR-defective phenotypes with alkylating agent tolerance and elevated mutation frequencies. Although, in these clones, microsatellites were not markedly destabilized as in tumors occurring in LS patients. The present finding suggested that, in addition to defective MMR, previously unrecognized molecular mechanisms may underlie repeat destabilization in human cells.
Case Study 3: Knockin
Reversible cloaking/uncloaking strategy with sgRNAs to control the CRISPR–Cas9 gene editing efficiency in vitro and in living cells
"RNA cloaking" is a mild and reversible chemical approach to controlling RNA hybridization, folding, and enzymatic interactions. In this study, the researcher used an azide-substituted acyl imidazole reagent (NAI-N3) to acylate the 20 -OH groups of RNAs post synthetically, resulting in blocking of the RNAs' folding, hybridization, and function. The activity of such poly-acylated (“cloaked”) RNAs in vitro was efficiently recovered upon treatment with a water-soluble phosphine that triggers Staudinger's reduction of the azide, followed by spontaneous loss of acyl groups (“uncloaking”). The researcher studies these reversible cloaking/uncloaking events with sgRNAs to control the CRISPR-Cas9 gene editing efficiency in vitro and in living cells. For the in-vitro study, they designed an in vitro gene editing model platform with a 103 nucleotide (nt) long synthetic sgRNA targeting a Cy5-labeled double-stranded DNA (dsDNA) target encoding a portion of the green fluorescent protein (GFP). They found that the highest levels of Cas9-mediated DNA cleavage -quantitatively complete restoration of activity to the level of untreated sgRNA-were obtained when the cloaked sgRNA was incubated with tris (hydroxypropyl) phosphine (THPP) or diphenylphosphinobenzene-3-sulfonate (TPPMS) in phosphate-buffered saline (PBS) buffer at 37 C for 1 hour at 1-5 mM concentrations. This strikingly robust performance in vitro directs the possible utility of this method in control of timing and initiation in diagnostic applications of CRISPR-Cas9. For in vivo study, the researcher used GFP-positive HeLa cells that were initially transfected with GFP-targeting sgRNA and Cas9 protein using nucleofection or commercial cationic lipids. They performed an experiment to test whether GFP-targeting sgRNAs that were cloaked based on the same procedure as in the in vitro experiments and delivered by CART were able to block CRISPR–Cas9 genome editing in the GFP-positive HeLa cells. They observed that the GFP-positive cells that were transfected with cloaked sgRNA/ Cas9 mRNA expressed the same level of GFP fluorescence as untreated cells, confirming that acylation of sgRNA blocks essentially all sgRNA activity in live cells. This study highlighted the utility of reversible RNA acylation as a novel method for temporal control of the genome-editing function.
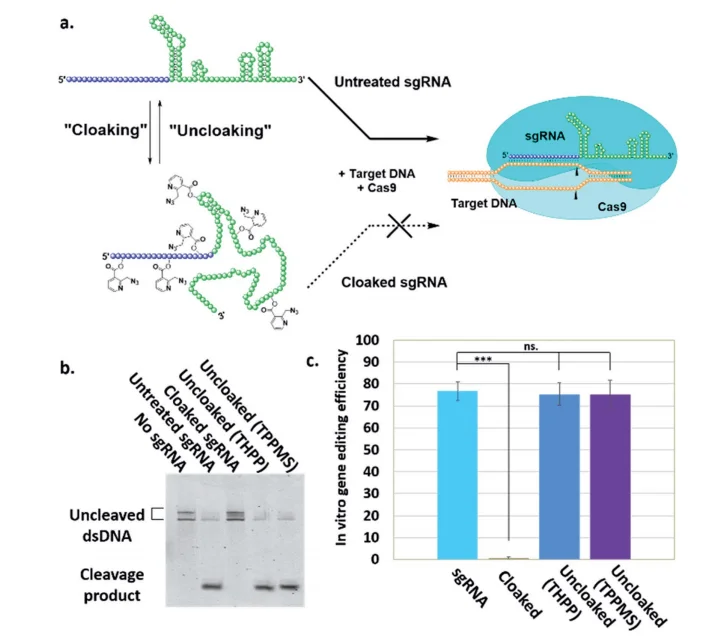
Fig 1: (a) Mechanism of NAI-N3-enabled inhibition of CRISPR–Cas9 gene editing. sgRNA cloaking inhibits the RNA-guided DNA double strand cleavage by Cas9 nuclease (b) PAGE analysis of Cas9 nuclease assay in vitro using Cy5-labelled dsDNA and untreated, cloaked, and uncloaked (phosphine-treated) sgRNA. (c) Bar chart represents the fraction of cleaved DNA after incubation of the sgRNAs with Cas9.
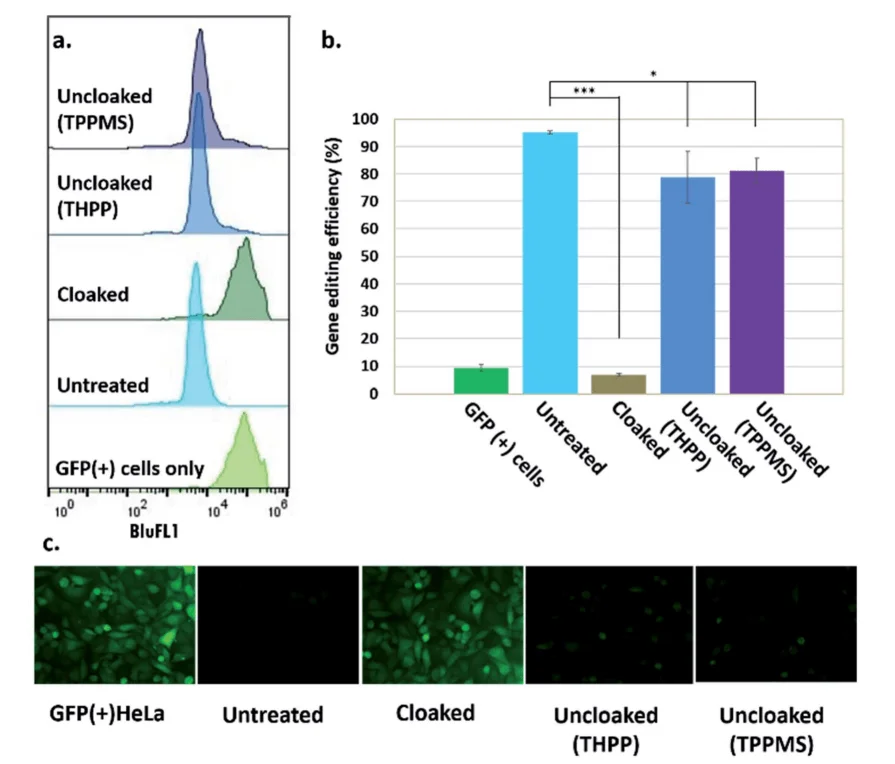
Fig 2: Phosphine control of cloaked CRISPR–Cas9 activity in human cells. (a) Flow cytometric analysis of GFP(+) HeLa cells showing loss of activity of cloaked sgRNA, and restoration of editing with phosphine treatment; (b) chart showing gene editing efficiencies (from flow cytometry data) of untreated, cloaked, and phosphine-uncloaked sgRNA (c) epifluorescence microscope images showing GFP knockout upon transfection with untreated sgRNA and Cas9 mRNA, lack of editing activity with cloaked sgRNA leaving GFP fluorescence unaffected, and restoration of editing with phosphine treatment of the cells.
Conclusion
With their unparalleled stability, rich genomic background, and long-standing contributions to scientific discovery, the HeLa cell line continues to be a powerful model for studying cancer, virology, drug response, and gene function. Whether used in fundamental research or advanced genome engineering, HeLa cells remain an essential tool for life scientists worldwide.
Ubigene provides high-quality, mycoplasma-free HeLa cells, STR-authenticated and registered in the ExPASy Cellosaurus database, ensuring traceability and experimental reliability. We also offer CRISPR-U™ gene editing services to help researchers create custom HeLa cell models — knockout, knockin, point mutation, or reporter tagged—for diverse research applications.
Looking to work with more trusted cells? >>>Explore our complete cell bank now!
Reference:
CRISPR/Cas9-Mediated TERT Disruption in Cancer Cells. Int. J. Mol. Sci. 2020, 21, 653.
Differential genomic de-stabilization in human cells with pathogenic MSH2 mutations introduced by genome editing. Experimental Cell Research, 2019 (377) 24–35.
Reversible RNA acylation for control of CRISPR-Cas9 gene editing. Chem. Sci., 2020, 11, 1011-1016.


