Ubigene Offers 4 Cutting-Edge Solutions for Point Mutation Cell Line Construction

Ubigene Offers 4 Cutting-Edge Solutions for Point Mutation Cell Line Construction

Background: The Rising Need for Precision SNP Models
The explosion of next-generation sequencing (NGS) has led to the discovery of millions of single nucleotide polymorphisms (SNPs), many of which are linked to human diseases such as cancer, neurodegenerative disorders, cardiovascular diseases, and inherited metabolic syndromes. To validate these SNPs functionally or develop precision therapies, researchers must construct cell lines with defined point mutations that accurately mimic disease genotypes.
However, traditional gene-editing methods like CRISPR/Cas9 often face low editing efficiency, off-target effects, and HDR limitations — particularly in complex or hard-to-edit loci.
Ubigene provides a comprehensive, experience-backed platform with four core methods, optimized to meet the diverse technical needs and biological constraints of SNP editing projects. Learn more>>
Method 1: RNP-Mediated Gene Editing - Fast and Reliable Point Mutation Strategy
Principle
The RNP (Ribonucleoprotein) approach utilizes a pre-assembled Cas9 protein and sgRNA complex, introduced directly into cells along with a single-stranded donor oligonucleotide (ssODN) containing the desired mutation. Cas9 creates a site-specific double-strand break, which is then repaired via homology-directed repair (HDR) using the ssODN template.
Experimental Workflow
1. Design sgRNA close to the target SNP (±20 bp)
2. Prepare Cas9 protein + sgRNA + ssODN (~120 nt)
3. Transfect cells via electroporation/lipofection
4. Validate edited clones by sequencing
✅ Advantages
1. High efficiency and rapid turnaround
2. No plasmid integration risk
3. Minimal off-target concerns due to transient presence of RNP
4. Suitable for a wide range of cell types
Limitations
1. Requires an optimal PAM site close to the SNP
2. RNA degradation and delivery efficiency can be challenges
3. Lower editing rate in non-dividing cells (HDR dependency)
Method 2: Prime Editing - Versatile Precision Editing Without DSB or Donor Template
Principle
Prime editing is a novel method combining Cas9 nickase (nCas9) fused to a reverse transcriptase, guided by a special prime editing guide RNA (pegRNA). This system directly writes new genetic information into the target locus—capable of all 12 base substitutions, insertions, or deletions—without inducing double-strand breaks or requiring a donor DNA.
Experimental Workflow
1. Design pegRNA: includes both target-binding and reverse transcription template
2. Deliver Prime Editor (nCas9-RT fusion protein) + pegRNA via plasmid or RNP
3. Perform nicking with additional sgRNA (PE3 system) to enhance efficiency
4. Sequence clones for accurate mutation detection
✅ Advantages
1. Capable of all base changes, insertions, deletions
2. No need for donor DNA or DSB
3. Lower indel formation than classical CRISPR
Limitations
1. Lower efficiency than base editing in some loci
2. Complex pegRNA design
3. Larger construct size (harder delivery for some cells)
4. Still under optimization for certain cell types
Method 3: Base Editing - High-Efficiency Single-Nucleotide Conversion Without DSB
Principle
Base editors consist of dCas9 or nCas9 fused with a DNA deaminase that enables direct base conversion (e.g., A→G or C→T) without breaking DNA strands or needing a repair template.
· ABE (Adenine Base Editor): A→G or T→C
· CBE (Cytosine Base Editor): C→T or G→A
· BE3 and ABE8e are widely used systems
Experimental Workflow
1. Design gRNA ensuring the target site falls within the editing window
2. Transfect plasmids encoding the base editor + gRNA
3. Screen edited clones by sequencing (PCR not sufficient)
✅ Advantages
1. Extremely high precision and editing efficiency
2. Avoids DSBs → lower indels and cell stress
3. Suitable for correction of pathogenic SNPs
Limitations
1. Restricted to specific transitions (not all base substitutions)
2. Editing window limitations and potential for off-target "bystander" mutations
3. Not suitable when target base is not unique in the window
Method 4: Antibiotics-Based Knock-In - Enhanced HDR via Selective Plasmid System
Principle
This method involves co-transfection of:
· A gRNA/Cas9 expression plasmid, and
· A donor plasmid containing long homology arms flanking a resistance gene cassette, inserted within an intron of the target gene. The resistance cassette is flanked by LoxP sites to allow subsequent removal via Cre recombinase.
Cells that have successfully undergone homology-directed recombination will survive antibiotic selection, enriching for positive clones.
Experimental Workflow
1. Design sgRNA at intronic regions near the SNP
2. Construct donor plasmid with 600-1000 bp homology arms + resistance cassette
3. Transfect cells and apply antibiotic selection
4. Transfect with Cre plasmid to remove resistance gene
5. Verify correct editing and removal by sequencing
✅ Advantages
1. Enhanced mutation efficiency due to antibiotic selection
2. Broader targetable regions (not limited by PAM proximity)
3. Especially useful when RNP or base editing is not feasible
Limitations
1. Longer experiment duration
2. Additional step required for Cre-mediated excision
3. Leaves LoxP scars in introns (minimized if well-positioned)
EZ-HRex™ – Ubigene’s Proprietary High-Efficiency HDR Platform
Ubigene has developed a powerful, patented HDR enhancement platform:
EZ-HRex™ Technology
Built upon the CRISPR-U™ system, EZ-HRex™ introduces the U+ Molecule to inhibit the non-homologous end joining (NHEJ) pathway, promoting precise HDR-based genome editing.
Key Benefits
1. Up to 84% HDR-positive rate in cell pools
2. Broadly applicable to SNP point mutations and gene knock-in
3. Compatible with RNP, plasmid, and AAV-based methods
4. Enhanced reproducibility and editing success
Learn More about the EZ-HRex™ Technology>>
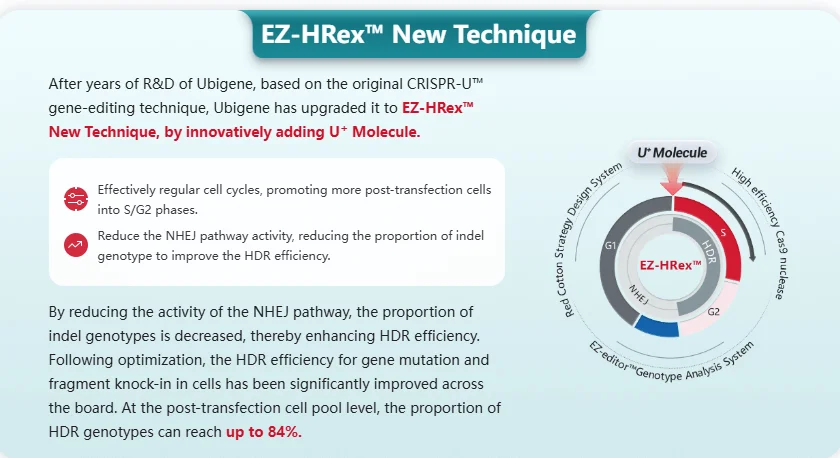
HDR enhancement platform - EZ-HRex™ Technology
Summary Comparison Table
| Method | Base Change Scope | DNA Break? | Donor Required | Efficiency | Best Use Case |
|---|---|---|---|---|---|
| RNP | Limited (via HDR) | Yes | ssODN | ★★★★☆ | Fast editing in common cell lines |
| Prime Editing | All base changes | No | No | ★★★☆☆ | Complex mutations, insertions, deletions |
| Base Editing | A→G / C→T only | No | No | ★★★★★ | Precise single-nucleotide change |
| Antibiotic Knock-in | Any via HDR | Yes | dsDonor (plasmid) | ★★★★☆ | Difficult loci, enhanced clone selection |
Need Expert Support? Let Ubigene Do the Work
Ubigene has successfully constructed CRISPR point mutation models in over 300 mammalian cell lines.
Special Promo:
As low as $4980 for point mutation cell line generation services. Request a quote>>
In-stock Cas9 Stable Expression Cell Lines Available: Just transfect gRNA + Donor to complete your edit with high efficiency.
Find your target cell line in our cell line bank>>

