Aspergillus nidulans, also known as Emericella nidulans, is one of the important fungal systems in genetics and cell biology. It has always been an important research organism for studying eukaryotic cell biology, including recombination, DNA repair, mutation, cell cycle control, tubulin, chromatin, lentiviral packaging, nuclear dynamics, pathogenicity, metabolism and experimental evolution.
Fungi play an important role in degrading the biomass in the ecosystem because they degrade all types of organic matter. Therefore, they are the main source of industry-related enzymes such as amylase, cellulase, lipase, pectinase and protease. The number of fully sequenced fungal genomes is rapidly increasing. Due to the poor development of genetic tools for most filamentous fungi, it is currently difficult to use genetic engineering to understand the biological characteristics of these fungi and make full use of them in industry. Therefore, in order to develop a universal method that can be used to genetically manipulate non-model filamentous fungi, scientists have developed a CRISPR-Cas9-based system suitable for filamentous fungi. CRISPR technology has revolutionized fungal genetic engineering by increasing the speed and complexity of experiments that can be performed. In addition, the efficiency of the system generally allows genetic engineering to be introduced into non-model species. Alfalfa (Emericella nidulans) has a long production history as a source of industrial chemicals and enzymes, and is a developmental model system for studying genetic regulation, developmental biology, signal transduction and secondary metabolism. Therefore, gene knockout, gene knock-in or point mutation will become a new method for studying Aspergillus nidulans.
Application of CRISPR in efficient marker-free gene targeting in Aspergillus nidulans
The survival of specific Cas9/sgRNA-mediated DNA double-strand breaks (DSB) depends on the addition of non-homologous ends to the NHEJ DNA repair pathway. We used this observation to develop a TAPE tool to evaluate the efficiency of the Aspergillus nidulans prototype spacer. Moreover, in NHEJ-deficient strains, effective marker-free gene targeting can be performed. Indeed, the study shows that even single-stranded oligonucleotides can be effectively used as a repair template for specific Cas9/sgRNA-induced DNA DSBs in Aspergillus nidulans and Aspergillus niger, indicating that this type of repair may be widely distributed in filamentous fungi Spreading. Importantly, by using single-stranded oligonucleotides for CRISPR-Cas9-mediated gene editing, specific point mutations and gene deletions (gene knockouts) can be introduced with an efficiency close to 100%. Therefore, two-point mutations and single gene insertion can be introduced very effectively in one transformation experiment.
Cpf1 can quickly and efficiently perform genome editing in Aspergilli
The efficiency of CRISPR gene editing is due to specific DNA double-strand breaks formed by RNA-guided nucleases. So far, among filamentous fungi, only Cas9 has been used as a CRISPR nuclease. Since genes edited with Cas9 are restricted by their 5'-NGG-3' protospacer adjacent motif (PAM) sequence, it is important to introduce RNA-guided nucleases that rely on other PAM sequences to target more Large genomic locus. The Cpf1 knockout of E. coli uses the PAM sequence composed of 5'-TTTN-3', so it is an attractive choice to achieve this goal. In this study, the Lb_cpf1 codon optimized for Aspergillus nidulans can be used for CRISPR-based gene editing in filamentous fungi. The researchers developed a vector-based setup for Cpf1-mediated CRISPR experiments and showed that it can work efficiently at different loci of Aspergillus nidulans and Aspergillus niger. Specifically, Cpf1 can catalyze oligonucleotide-mediated genome-directed mutagenesis and marker-free gene targeting. The results show that Cpf1 can be effectively used for gene editing in Aspergillus, thereby expanding the range of genomic DNA sequences that can be targeted by CRISPR technology.
Efficient CRISPR knockout in Nidulans
In Aspergillus nidulans, gene targeting can be performed by homologous recombination in the transformation process, but the frequency of correct gene targeting is variable and usually low. The researchers identified Aspergillus nidulans (nkuA) of the human KU70 gene, which is essential for non-homologous end joining of DNA in double-strand break repair. The deletion of nkuA (nkuAΔ) greatly reduces the frequency of non-homologous integration of the transformed DNA fragments, thereby significantly improving gene targeting. Alternative heterologous markers have also been developed in Aspergillus nidulans, but integration is not guided at any time in the Aspergillus nidulans genome. Together, nkuAΔ and heterologous selection markers form a very effective gene targeting sy.
The efficiency of gene knock-out and cleavage can not only give people the ability to generate protein radical profiles and establish regulatory records, but also has many advantages, making it a particularly attractive recombinant protein expression system. First, it is carboxylated on glutamic acid and sulfated on tyrosine. Second, the operation is simple, and the recombinant protein can be quickly produced through transient gene expression. Third, it can be used for stable recombinant protein production. Some researchers used gene cell knockout and cutting efficiency systems to generate gene-edited cell lines, targeted sequencing of GLUL genomic loci, produced stable transgenic EPO cell lines, and discovered the mechanism of stable expression of recombinant erythropoietin in humans .
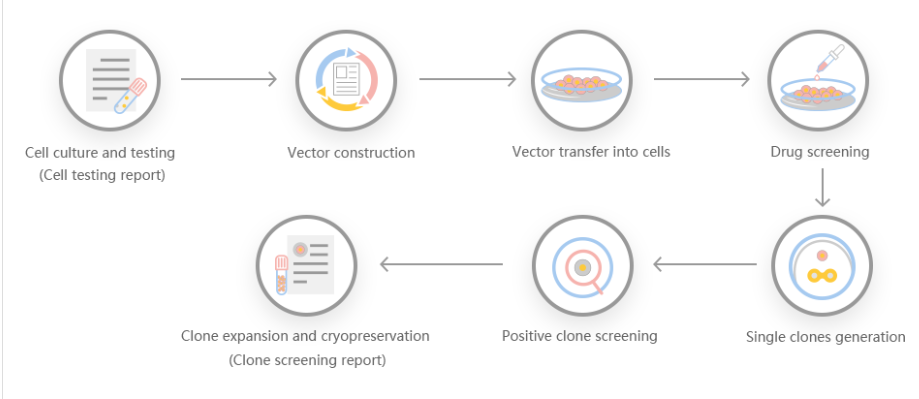
According to customer needs, Yuanjing Biotechnology designs a stable gene transfer knockout program based on the target gene.
Scheme 1: Small-segment gene knockout scheme, gRNA is set in the introns at both ends of exon 2, and the number of coding bases of the knockout exon is not 3 times, and the knockout can cause frame shift.
Scheme 2: Frameshift gene knockout scheme, gRNA is set on the exon, the number of missing bases is not 3 times, and frameshift mutation can occur after knockout.
Scheme 3: Large-segment gene knockout scheme, knock out the coding sequence of the entire gene to achieve the effect of large-segment knockout.
Ubigene Biosciences is co-founded by biological academics and elites from China, the United States, and France. We are located in Guangzhou Science City, which serves as a global center for high technology and innovation. Ubigene Biosciences has 1000㎡ office areas and laboratories, involving genome editing, cell biology technology, and zebrafish research. We provide products and services for plasmids, viruses, cells, and zebrafish. We aim to provide customers with better gene-editing tools for cell or animal research.
We developed CRISPR-U™ and CRISPR-B™(based on CRISPR/Cas9 technology) which is more efficient than general CRISPR/Cas9 in double-strand breaking, CRISPR-U™ and CRISPR-B™ can greatly improve the efficiency of homologous recombination, easily achieve knockout (KO), point mutation (PM) and knockin (KI) in vitro and in vivo.
References:
1. Nødvig CS, Hoof JB, Kogle ME, et al. Efficient oligonucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet Biol. 2018;115:78-89.
2. Nødvig CS, Nielsen JB, Kogle ME, Mortensen, UH. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS One. 2015;10(7):e0133085. Published 2015 Jul 15.
3. Vanegas, K.G., Jarczynska, Z.D., Strucko, T. et al. Cpf1 enables fast and efficient genome editing in Aspergilli. Fungal Biol Biotechnol 6, 6 (2019).
4. Nayak T, Szewczyk E, Oakley CE, et al. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172(3):1557-1566.
Genome Editing Platform
——Focusing on the Application of CRISPR-U™ and CRISPR-B™ Gene Editing Technology
1. Provides various types of gene-editing vectors for different species.
2. Provides different virus packaging services, including lentiviruses, adenoviruses and adeno-associated viruses.3. Provides high-quality services for gene knockout, point mutation and knockin cell lines.
Cell Biology Platform
——Focusing on primary cell
1. Provides over 400 types of primary cells.
2. Provides culture strategies and related products for different cell types.3. Provides cell biology-related services such as cell isolation, extraction and validation.
 Subscribe Us
Subscribe Us Gene Editing Services
Gene Editing Services
 EZ-editor™
EZ-editor™ Red Cotton Gene knockout Project
Red Cotton Gene knockout Project








