Saccharomyces cerevisiae is a species of yeast. It has been instrumental to winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes (one can see the yeast as a component of the thin white film on the skins of some dark-colored fruits such as plums; it exists among the waxes of the cuticle). It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like Escherichia coli as the model bacterium. It is the microorganism behind the most common type of fermentation.
Phosphoglycerate mutase knock-out mutant Saccharomyces cerevisiae: physiological investigation and transcriptome analysis
The yeast Saccharomyces cerevisiae is able to adapt its metabolism to grow on different carbon sources and to shift to non-fermentative growth on C2 or C3 carbon sources (ethanol, acetate, or glycerol) through the activation of gluconeogenesis. Researcher studied the response to the deletion of the glycolytic and gluconeogenic gene GPM1, encoding for phosphoglycerate mutase. It was previously shown that a S. cerevisiae strain with non-functional copies of GPM1 can only grow when glycerol and ethanol are both present as carbon sources, whilst addition of glucose was shown to strongly inhibit growth. It was suggested that glycerol is needed to feed gluconeogenesis whilst ethanol is required for respiration. Researcher studied the physiological response of the GPM1 knock-out mutant through fermentation and transcriptome analysis. Furthermore, researcher compared the physiological results with those obtained through simulations using a genome-scale metabolic model, showing that glycerol is only needed in small amounts for growth. Our findings strongly suggest a severely impaired growth ability of the knock-out mutant, which presents increased transcript levels of genes involved in the pentose phosphate pathway and in the glyoxylate shunt. These results indicate an attempt to compensate for the energy imbalance caused by the deletion of the glycolytic/gluconeogenic gene within the mutant.
Reducing phenolic off-flavors through CRISPR-based gene editing of the FDC1 gene in Saccharomyces cerevisiae x Saccharomyces eubayanus hybrid lager beer yeasts
Today’s beer market is challenged by a decreasing consumption of traditional beer styles and an increasing consumption of specialty beers. In particular, lager-type beers (pilsner), characterized by their refreshing and unique aroma and taste, yet very uniform, struggle with their sales. The development of novel variants of the common lager yeast, the inter-specific hybrid Saccharomyces pastorianus, has been proposed as a possible solution to address the need of product diversification in lager beers. Previous efforts to generate new lager yeasts through hybridization of the ancestral parental species (S. cerevisiae and S. eubayanus) yielded strains with an aromatic profile distinct from the natural biodi-versity. Unfortunately, next to the desired properties, these novel yeasts also inherited unwanted characteristics. Most notably is their phenolic off-flavor (POF) production, which hampers their direct application in the industrial production processes. Researchers describe a CRISPR-based gene editing strategy that allows the systematic and meticulous intro-duction of a natural occurring mutation in the FDC1 gene of genetically complex industrial S. cerevisiae strains, S. eubayanus yeasts and interspecific hybrids. The resulting cis-genic POF- variants show great potential for industrial application and diversifying the cur-rent lager beer portfolio.
A CRISPR/Cas9-based exploration into the elusive mechanism for lactate export in Saccharomyces cerevisiae.
CRISPR/Cas9-based genome editing allows rapid, simultaneous modification of multiple genetic loci in Saccharomyces cerevisiae. This technique was used in a functional analysis study aimed at identifying the hitherto unknown mechanism of lactate export in this yeast. First, an S. cerevisiae strain was constructed with deletions in 25 genes encoding transport proteins, including the complete aqua(glycero)porin family and all known carboxylic acid transporters. The 25-deletion strain was then transformed with an expression cassette for Lactobacillus casei lactate dehydrogenase (LcLDH). In anaerobic, glucose-grown batch cultures this strain exhibited a lower specific growth rate (0.15 vs. 0.25 h−1) and biomass-specific lactate production rate (0.7 vs. 2.4 mmol g biomass−1 h−1) than an LcLDH-expressing reference strain. However, a comparison of the two strains in anaerobic glucose-limited chemostat cultures (dilution rate 0.10 h−1) showed identical lactate production rates. These results indicate that, although deletion of the 25 transporter genes affected the maximum specific growth rate, it did not impact lactate export rates when analysed at a fixed specific growth rate. The 25-deletion strain provides a first step towards a ‘minimal transportome’ yeast platform, which can be applied for functional analysis of specific (heterologous) transport proteins as well as for evaluation of metabolic engineering strategies.
The efficiency of gene knock-out and cleavage can not only give people the ability to generate protein radical profiles and establish regulatory records, but also has many advantages, making it a particularly attractive recombinant protein expression system. First, it is carboxylated on glutamic acid and sulfated on tyrosine. Second, the operation is simple, and the recombinant protein can be quickly produced through transient gene expression. Third, it can be used for stable recombinant protein production. Some researchers used gene cell knockout and cutting efficiency systems to generate gene-edited cell lines, targeted sequencing of GLUL genomic loci, produced stable cell lines, and discovered the mechanism of stable expression of recombinant erythropoietin in humans .
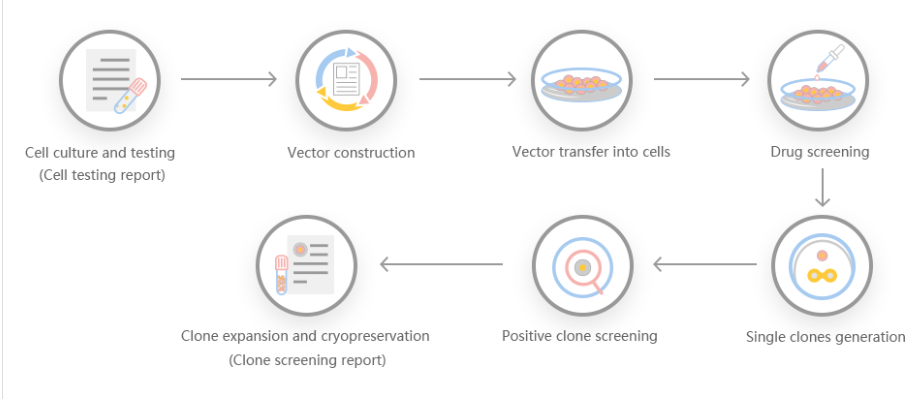
According to customer needs, Yuanjing Biotechnology designs a stable gene transfer knockout program based on the target gene.
Scheme 1: Small-segment gene knockout program, gRNA is set in the introns at both ends of exon 2, and the number of bases encoded by the knockout exon is not 3 times, and the knockout can cause frameshift.
Scheme 2: Frameshift gene knockout scheme, gRNA is set on the exon, the number of missing bases is not 3 times, and frameshift mutation can occur after knockout.
Scheme 3: Large-segment gene knockout scheme, knock out the coding sequence of the entire gene to achieve the effect of large-segment knockout.
Reference
Papini M , Nookaew I , Scalcinati G , et al. Phosphoglycerate mutase knock-out mutant Saccharomyces cerevisiae: Physiological investigation and transcriptome analysis[J]. Biotechnology Journal, 2010, 5(10):1016-1027.
Mertens S , Gallone B , Steensels J , et al. Reducing phenolic off-flavors through CRISPR-based gene editing of the FDC1 gene in Saccharomyces cerevisiae x Saccharomyces eubayanus hybrid lager beer yeasts[J]. Plos One, 2019, 14(1).
Robert, Mans, Else-Jasmijn, et al. A CRISPR/Cas9-based exploration into the elusive mechanism for lactate export in Saccharomyces cerevisiae.[J]. FEMS yeast research, 2017.
Ubigene Biosciences is co-founded by biological academics and elites from China, the United States, and France. We are located in Guangzhou Science City, which serves as a global center for high technology and innovation. Ubigene Biosciences has 1000㎡ office areas and laboratories, involving genome editing, cell biology technology, and zebrafish research. We provide products and services for plasmids, viruses, cells, and zebrafish. We aim to provide customers with better gene-editing tools for cell or animal research.
We developed CRISPR-U™ and CRISPR-B™(based on CRISPR/Cas9 technology) which is more efficient than general CRISPR/Cas9 in double-strand breaking, CRISPR-U™ and CRISPR-B™ can greatly improve the efficiency of homologous recombination, easily achieve knockout (KO), point mutation (PM) and knockin (KI) in vitro and in vivo.
Genome Editing Platform
——Focusing on the Application of CRISPR-U™ and CRISPR-B™ Gene Editing Technology
1. Provides various types of gene-editing vectors for different species.
2. Provides different virus packaging services, including lentiviruses, adenoviruses and adeno-associated viruses.3. Provides high-quality services for gene knockout, point mutation and knockin cell lines.
Cell Biology Platform
——Focusing on primary cell
1. Provides over 400 types of primary cells.
2. Provides culture strategies and related products for different cell types.3. Provides cell biology-related services such as cell isolation, extraction and validation.
 Subscribe Us
Subscribe Us Gene Editing Services
Gene Editing Services
 EZ-editor™
EZ-editor™ Red Cotton Gene knockout Project
Red Cotton Gene knockout Project








