Genes with sequence similarity to the yeast tRNA (His) guanosine transferase (Thg1) gene have been identified in all three domains of life. The Thg1 family of enzymes involves a variety of processes from tRNA (His) maturation to 5'end repair, such as knockout cell line. tRNA. All of these activities take advantage of the ability of the Thg1 family of enzymes to catalyze 3'-5' nucleotide addition reactions. Although many Thg1-containing organisms have only one Thg1-related gene, some eukaryotic microorganisms have multiple genes similar in sequence to Thg1.
Structural study of bacterial tRNAHIS guanosyltransferase (Thg1)-like protein with nucleotides in activation and nucleotide transfer sites
All nucleotide polymerases and transferases catalyze the addition of nucleotides in the 5'to 3'direction. In contrast, the tRNAHis guanidinotransferase (Thg1) enzyme catalyzes the abnormal addition (3' to 5') of nucleotides to polynucleotide substrates. In eukaryotes, Thg1 enzyme uses 3'-5' additional activity to add G-1 to the 5'end of tRNAHis, which is a necessary modification for histidine-tRNA synthetase to effectively aminoacylate tRNA. Thg1-like protein (TLP) is found in archaea, bacteria and mitochondria, and is biochemically different from its eukaryotic Thg1 counterpart, TLP, which catalyzes the repair of the 5'end of truncated tRNA and acts on a wide range of tRNA substrates. It does not exhibit strictly specific tRNAHis. Taken together, these data of knockout bacteria indicate that TLPs play a role in different biological pathways of tRNAHis maturation pathway, perhaps in tRNA quality control. The researchers introduced the first crystal structure of TLP from the Gram-positive soil bacterium Bacillus thuringiensis (BtTLP). The enzyme is a tetramer similar to human THG1, and has basic structural similarities with it. Catalyzing the 3'-5' reaction with 5'-monophosphorylated tRNA first requires an activation step, that is, the 5'-adenylation intermediate is generated before the second nucleotide base transfer step, in which the nucleotide is Transfer to the 5'-end of the tRNA. Consistent with the early characteristics of human THG1, the researchers observed unique binding sites for nucleotides involved in the two steps of activation and nucleotide transfer. The BtTLP complex with GTP reveals a new interaction with the GTP nucleotide at the activation site, which is invisible from the previously resolved structure. In addition, the BtTLP-ATP structure allows direct observation of ATP at the activation site for the first time. The BtTLP structure data, combined with the kinetic analysis of selected variants, provide new insights into the role of key residues in the activation step.
Saccharomyces cerevisiae Thg1 uses 5'-pyrophosphate removal to control addition of nucleotides to tRNA(His.)
In eukaryotes, the tRNA(His) guanylyltransferase (Thg1) catalyzes 3'-5' addition of a single guanosine residue to the -1 position (G-1) of tRNA(His), across from a highly conserved adenosine at position 73 (A73). After addition of G-1, Thg1 removes pyrophosphate from the tRNA 5'-end, generating 5'-monophosphorylated G-1-containing tRNA. The presence of the 5'-monophosphorylated G-1 residue is important for recognition of tRNA(His) by its cognate histidyl-tRNA synthetase. In addition to the single-G-1 addition reaction, Thg1 polymerizes multiple G residues to the 5'-end of tRNA(His) variants. For 3'-5' polymerization, Thg1 uses the 3'-end of the tRNA(His) acceptor stem as a template. The mechanism of reverse polymerization is presumed to involve nucleophilic attack of the 3'-OH from each incoming NTP on the intact 5'-triphosphate created by the preceding nucleotide addition. The potential exists for competition between 5'-pyrophosphate removal and 3'-5' polymerase reactions that could define the outcome of Thg1-catalyzed addition, yet the interplay between these competing reactions has not been investigated for any Thg1 enzyme. Researcher establish transient kinetic assays to characterize the pyrophosphate removal versus nucleotide addition activities of yeast Thg1 with a set of tRNA(His) substrates in which the identity of the N-1:N73 base pair was varied to mimic various products of the N-1 addition reaction catalyzed by Thg1. Researcher demonstrate that retention of the 5'-triphosphate is correlated with efficient 3'-5' reverse polymerization. A kinetic partitioning mechanism that acts to prevent addition of nucleotides beyond the -1 position with wild-type tRNA(His) is proposed.
Presence of a classical RRM-fold palm domain in Thg1-type 3'- 5'nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains
Using sensitive profile-profile comparison and structure prediction methods we show that the catalytic domain Thg1 contains a RRM (ferredoxin) fold palm domain, just like the viral RNA-dependent RNA polymerases, reverse transcriptases, family A and B DNA polymerases, adenylyl cyclases, diguanylate cyclases (GGDEF domain) and the predicted polymerase of the CRISPR system. Researcher show just as in these polymerases, Thg1 possesses an active site with three acidic residues that chelate Mg++ cations. Based on this researcher predict that Thg1 catalyzes polymerization similarly to the 5'-3' polymerases, but uses the incoming 3' OH to attack the 5' triphosphate generated at the end of the elongating polynucleotide. In addition researcher identify a distinct set of residues unique to Thg1 that researcher predict as comprising a second active site, which catalyzes the initial adenylation reaction to prime 3'-5' polymerization. Based on contextual information from conserved gene neighborhoods researcher show that Thg1 might function in conjunction with a polynucleotide kinase that generates an initial 5' phosphate substrate for it at the end of a RNA molecule. In addition to histidinyl tRNA maturation, Thg1 might have other RNA repair roles in representatives from all the three superkingdoms of life as well as certain large DNA viruses. Researcher also present evidence that among the polymerase-like domains Thg1 is most closely related to the catalytic domains of the GGDEF and CRISPR polymerase proteins.
Based on this relationship and the phyletic patterns of these enzymes we infer that the Thg1 protein is likely to represent an archaeo-eukaryotic branch of the same clade of proteins that gave rise to the mobile CRISPR polymerases and in bacteria spawned the GGDEF domains. Thg1 is likely to be close to the ancestral version of this family of enzymes that might have played a role in RNA repair in the last universal common ancestor.
Ubigene developed CRISPR-B™ which optimizes the microbial gene-editing vectors and process. The efficiency and accuracy are much higher than traditional methods. CRISPR-B™ can be used in gene editing of bacteria and fungi.
Reference
Song Z, Yu H, Wang P, et al. Germ cell-specific Atg7 knockout results in primary ovarian insufficiency in female mice.[J]. Cell Death and Disease, 2015, 6(1).
Da Silva AM, Maciel RM, Da Silva MR, Toledo SR, De Carvalho MB, Cerutti JM. A novel germ-line point mutation in RET exon 8 (Gly(533)Cys) in a large kindred with familial medullary thyroid carcinoma. J Clin Endocrinol Metab. 2003;88(11):5438-5443. doi:10.1210/jc.2003-030997
ZhigangYu,NahidDadgar,MeganAlbertelli,et al.Abnormalities of Germ Cell Maturation and Sertoli Cell Cytoskeleton in Androgen Receptor 113 CAG Knock-In Mice Reveal Toxic Effects of the Mutant Protein.American Journal of Pathology,2006. 195-203.
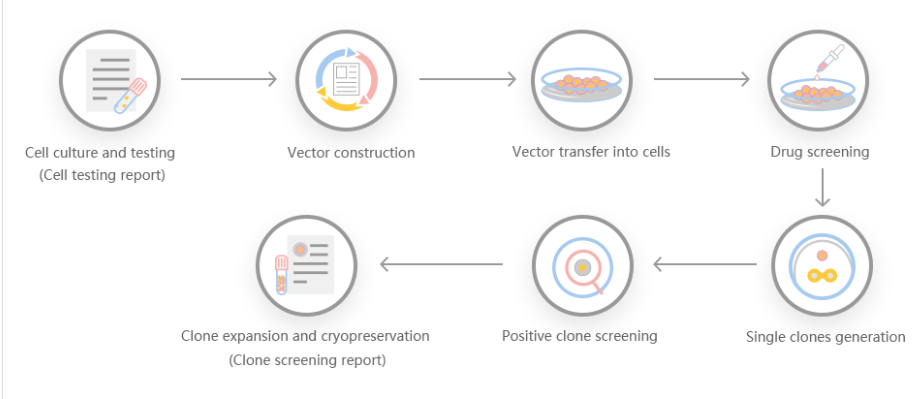
The efficiency of gene knock-out and cleavage can not only give people the ability to generate protein radical profiles and establish regulatory records, but also has many advantages, making it a particularly attractive recombinant protein expression system. First, it is carboxylated on glutamic acid and sulfated on tyrosine. Second, the operation is simple, and the recombinant protein can be quickly produced through transient gene expression. Third, it can be used for stable recombinant protein production. Some researchers used gene cell knockout and cutting efficiency systems to generate gene-edited cell lines, targeted sequencing of GLUL genomic loci, produced stable EPO cell lines, and discovered the mechanism of stable expression of recombinant erythropoietin in humans .
According to customer needs, Yuanjing Biotechnology designs a stable gene transfer knockout program based on the target gene
Scheme 1: Small-segment gene knockout program, gRNA is set in the introns at both ends of exon 2, and the number of bases encoded by the knockout exon is not 3 times, and the knockout can cause frameshift.
Scheme 2: Frameshift gene knockout scheme, gRNA is set on the exon, the number of missing bases is not 3 times, and frameshift mutation can occur after knockout.
Scheme 3: Large-segment gene knockout scheme, knock out the coding sequence of the entire gene to achieve the effect of large-segment knockout.
Ubigene Biosciences is co-founded by biological academics and elites from China, the United States, and France. We are located in Guangzhou Science City, which serves as a global center for high technology and innovation. Ubigene Biosciences has 1000㎡ office areas and laboratories, involving genome editing, cell biology technology, and zebrafish research. We provide products and services for plasmids, viruses, cells, and zebrafish. We aim to provide customers with better gene-editing tools for cell or animal research.
We developed CRISPR-U™ and CRISPR-B™(based on CRISPR/Cas9 technology) which is more efficient than general CRISPR/Cas9 in double-strand breaking, CRISPR-U™ and CRISPR-B™ can greatly improve the efficiency of homologous recombination, easily achieve knockout (KO), point mutation (PM) and knockin (KI) in vitro and in vivo.
Genome Editing Platform
——Focusing on the Application of CRISPR-U™ and CRISPR-B™ Gene Editing Technology
1. Provides various types of gene-editing vectors for different species.
2. Provides different virus packaging services, including lentiviruses, adenoviruses and adeno-associated viruses.3. Provides high-quality services for gene knockout, point mutation and knockin cell lines.
Cell Biology Platform
——Focusing on primary cell
1. Provides over 400 types of primary cells.
2. Provides culture strategies and related products for different cell types.3. Provides cell biology-related services such as cell isolation, extraction and validation.
 Subscribe Us
Subscribe Us Gene Editing Services
Gene Editing Services
 EZ-editor™
EZ-editor™ Red Cotton Gene knockout Project
Red Cotton Gene knockout Project








