ncRNA is
closely relating to disease, so how can we study its function?
The sequencing
results of the human genome show that there are more than 20,000 genes
responsible for encoding proteins, accounting for less than 2% of the total genome
length. Many protozoa also have more than 20000 protein coding genes, but
unlike humans, the proportion of coding genes in these creatures in the total
gene length is much higher. Therefore, interesting questions came out: Does the
98% non-coding RNA (ncRNA) contribute to the complexity of the human body? How
does ncRNA participate in life activities?


Figure 1. Proportion of ncRNAs in different species
At first, people
had limited knowledge of ncRNA, and believed that ncRNAs are junk genes, which
might be just a "shelter" to avoid external adverse environments,
such as rays, viruses, and other damage to human genes. With the deepening of
research, scientists gradually discovered the importance of ncRNAs. ncRNAs do
not encode proteins, but they are not non-transcribed. In fact, these ncRNAs
are also continuously transcribed to participate in the protein synthesis
process. Among them, the most familiar one is tRNA that transport amino acids,
rRNA and snoRNA that participate in ribosome synthesis. At present, lncRNA,
microRNA and circRNA are the three main types highly related to cancer, which
play important roles in cancer cell proliferation, migration, cell death
resistance, etc. In clinical cancer screening, ncRNAs have also become an
important hallmark, providing great help for early screening of cancer. [1-4]

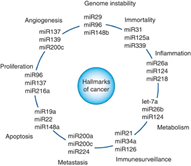

Figure 2. ncRNAs have become hallmarks of cancer
microRNA is referred to as miRNA, which
is complementary to mRNA through the principle of base pairing, leading to mRNA
cleavage and degradation. Although the principle of miRNA is single, it plays a
regulatory role in all aspects of cancer development. If miRNA can combine with
the mRNA of tumor suppressor genes, miRNA can promote tumor survival and proliferation.
On the contrary, if it is bound to the mRNA of oncogenes, miRNA can inhibit the
expansion of tumor cells.
The functions of lncRNA are more diverse: ① lncRNA directly binds
with mRNA to inhibit its translation, or combines with protein to form a
transcription regulatory complex, which acts as a transcription regulatory
factor to regulate the transcription level of downstream genes; ② It acts as a miRNA sponge, specifically binds to some miRNA, and
continuously interferes with the function of miRNA; ③ As the backbone of the intracellular complex, it enables the stable combination
of proteins and RNAs.
circRNA is lncRNA in essence. This long
chain non coding RNA will form a single chain closed loop after being
transcribed. Its mechanism of action is similar to that of IncRNA, which can
regulate the transcription of oncogenes and tumor suppressor genes, and can also
act as a molecular sponge to competitively inhibit miRNA. It has been suggested
that it can act as a molecular scaffold to provide a binding site for complexes.
Functional study methods and knockout
strategies of ncRNAs
To study the
function of ncRNA, it is necessary to edit its genes. RNAi technology is a
convenient and commonly used means in scientific research. However, due to its
low efficiency and off-target possibility, better regulation methods are
required. Compared with RNAi technology, CRISPR/Cas9 system has the advantages
of low off-target effect, more thorough knockout, and the ability to interfere
with RNA in the nucleus that is difficult to interfere with RNAi. CRISPR/Cas9
is more suitable for ncRNA intervention and gene knockout.5,6 The
following are CRISPR/Cas9 knockout strategies for miRNA, lncRNA, and circRNA:
The miRNA
fragment is small, so it is relatively simple to knockout. The target sequence
can be designed on the mature miRNA, at the stem ring. Or design a pair of
gRNAs to directly knockout the entire miRNA sequence, but it should be noted
that the knockout region cannot overlap with other coding genes.
LncRNA is
relatively long, and it is difficult to have a great impact on the function by
knockout some bases, so it is recommended to knockout as long fragment as
possible. Generally, large fragment gene knockout strategy is used, which is to
design 2-4 gRNAs from upstream to downstream.
However, circRNAs often overlap with the exons of coding genes, so the
knockout is more difficult and complex - it is necessary to avoid destroying
the exons while knockout circRNA. It is recommended to knockout the circRNA
loop forming element located in the intron, which can destroy the function of
circRNA without affecting the coding gene (Figure 3).

Figure 3. circHIPK3 knockout strategy - Alu looping
element knockout [8]
Ubigene
has developed CRISPR-U™ system, which is
10 times more efficient in editing. And Ubigene has mastered three knockout strategies:
small fragment knockout, indel frameshift mutation, and large fragment knockout.
More than 5000 successful cases of gene editing in 200+ cell lines. Easily achieve
the knockout of miRNA, lncRNA, and circRNA. Express knockout cell line service
as fast as four weeks, deliver high-quality positive clones. To learn more>>
After understanding the roles of ncRNA in tumor and the strategies
of ncRNA knockout, let's go over some cases of CRISPR/Cas9 mediated ncRNA
research.
Study of drug resistance
mechanism of tumor by CRISPR/Cas9 knockout miR-137
In
the article microRNA-137 promotes apoptosis in ovarian cancer cells via
the regulation of XIAP7 published by the British Journal
of Cancer, Jiang’s team from the State Key Laboratory of Biocontrol, School
of Life Sciences, Sun Yat-sen University clarified that miR-137 plays an
important role in cisplatin induced apoptosis of ovarian cancer cells.[7]
At present, it can be confirmed that
cisplatin can achieve the therapeutic effect by promoting tumor cells to
apoptosis, but its mechanism is still unclear. To uncover its mechanism,
Jiang's team screened eight miRNAs that can regulate apoptosis regulator XIAP (the
most effective apoptosis regulator in mammalian cells) through the dual
luciferase reporter gene. Among them, miR-137 has high significance, and the
high expression of this miR-137 also plays an important role in inhibiting
other tumors. Therefore, the team chose miR-137 for further study. By
overexpressing miR-137, ovarian cancer cell A2780 downregulates apoptosis
regulator XIAP, making ovarian cancer cells more prone to apoptosis when
induced by cisplatin.

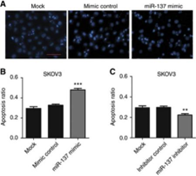
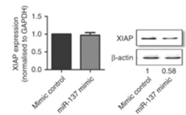
Figure 4. miR-137 promotes apoptosis of ovarian cancer
cells
In order to further confirm that miR-137
plays the role of tumor suppressor gene in the apoptosis of ovarian cancer
cells, Jiang's team designed three types of sgRNAs to target the upstream,
downstream and precise regions of mature miR-137, and knockout the miR-137 in
A2780 cell line using CRISPR/Cas9 system. It was found that after miR-137 was deleted,
XIAP was subsequently up-regulated, and the apoptosis rate was reduced.
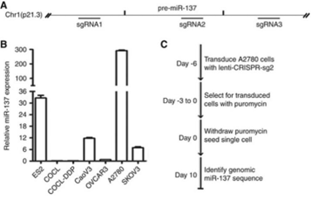
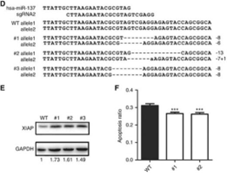
Figure 5. After knockout miR-137, XIAP is subsequently up-regulated
Study of the mechanism of tumor cell
proliferation and colony forming ability by CRISPR/Cas9 knockout SNHG1
Juan's team from
Center for Computational and Systems Biology of National Taiwan University
found that the high expression of SNHG1 was highly correlated with the poor
prognosis of neuroblastoma (NB). So Juan's team designed two gRNAs and knockout
the endogenous SNHG1 of SK-N-BE(2)C cell line (a NB cell line) using
CRISPR/Cas9 technology. In the article LncRNA SNHG1 regulates
neuroblastoma cell fate via interactions with HDAC1/28, Juan's team suggested that the knockout of SNHG1 inhibited the
proliferation of NB cells. Moreover, the analysis of cell line transcriptome
suggests that SNHG1 is closely related to a variety of biological processes,
such as cell migration, growth and apoptosis resistance. Moreover, SNHG1 can
affect the expression of genes in the core regulatory circuit (CRC), which
includes PHOX2B, HAND2, GATA3, ISL1, TBX1 and MYCN.
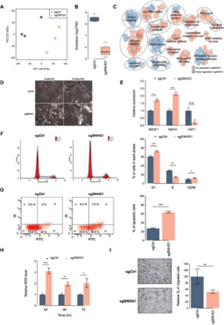
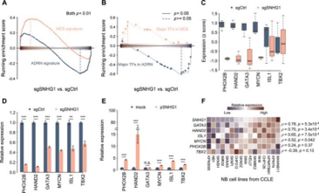
Figure 6. SNHG1 affects the core regulation loop
Juan’s team further used ChIP-seq and ATAC-seq
to study the possible interaction between SNHG1 and HDAC1/2, and then further
confirmed the interaction through pull-down and RIP.
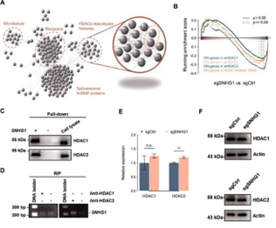
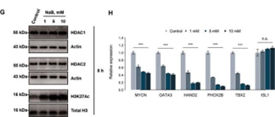
Figure 7. Results of pull-down and RIP
The above Juan’s team analyzed the action
mechanism of SNHG1 in NB, and suggested that blocking the interaction between
SNHG1 and HDAC1/2 would be essential to inhibit the proliferation and colony
formation of NB cells, providing a potential therapeutic strategy for the
treatment of NB.
Study the signal pathway of hepatocellular carcinoma development by
CRISPR/Cas9 technology
Compared
with normal liver tissues, circSOD2 is highly expressed in tumor tissues of HCC
patients. Inhibition of circSOD2 can significantly inhibit the proliferation of
liver cancer cells and slow down the tumor formation in vivo. The Tu’s team of
Wenzhou Medical University proposed a new signal pathway in the CircSOD2
induced epigenetic alteration drives hepatocellular carcinoma progression
through activating JAK2/STAT3 signaling pathway published in the Journal of Experimental & Clinical Cancer Research, which
provides new ideas for the prevention and drug intervention of liver cancer.
Tu’s
team analyzed the RNA interactome database, used Ago2 pull-down and biotin pull-down
techniques, and observed the expression level of miR-502-5p after silencing and
overexpressing CircSOD2. They found that CircSOD2 acts as a "miRNA
sponge" in liver tumor cells, specifically binding to miR-502-5p,
effectively inhibiting the expression of miR-502-5p.
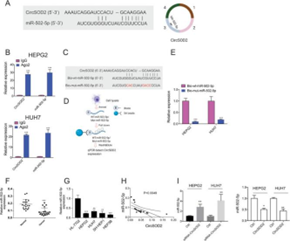
Figure 8. CircSOD2 inhibits the expression of
miR-502-5p
Subsequently,
Tu’s team made use of the above similar experimental principle, and found that
after adding the analog of miR-502-5p, the expression of DNMT3a, a protein
highly expressed in various liver cancer cell lines, was significantly down
regulated.
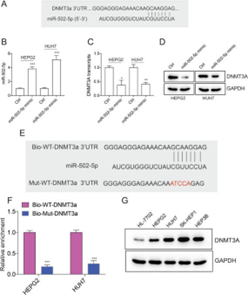
Figure 9. Down-regulation of DNMT3a by miR-502-5p
In
order to further study the molecular mechanism of DNMT3a in HCC, the team
constructed DNMT3a-KO HEPG2 and HUH7 cell lines by CRISPR/Cas9 technology. The
WB results show that DNMT3a may regulate the proliferation of HCC cells through
SOCS3/JAK2/STAT3 signal pathway. According to the clue that DNMT3a is a DNA
methyltransferase, the team further verified the reliability of the signal
pathway by using the dual luciferase reporter gene experiment and CHIP-qPCR
experiment. The final conclusion is that DNMT3a upregulation promotes SOCS3
promoter methylation and inhibits SOCS3 expression, thereby further activating
JAK2/STAT3 signal pathway.

Figure 10. DNMT3a inhibits SOCS3 expression, thereby
activates JAK2/STAT3 signaling pathway
Summary
Many ncRNAs have
been proved to play important roles in promoting tumor growth, migration and
escaping cell death. Meanwhile, some other ncRNAs play the role of tumor
suppressor, prevent cells from entering the division cycle, activate programmed
death, maintain genomic stability, and directly or indirectly inhibit tumor
occurrence.
With the help of CRISPR/Cas9 gene editing
technology, the above cases have achieved ideal results in the functional
research of ncRNAs. Ubigene has collected 200+ cell lines gene editing
parameters, and developed CRISPR-UTM system with 10X efficiency,
which can provide services for lncRNA, miRNA, and circRNA knockout in various cell
lines. High-quality homozygous KO clones deliver as fast as 4 weeks!
There are 3000+ in-stock KO cell lines available,
as low as $1980, deliver in 1 week. To explore your ideal KO cell line>>
Reference:
Winkle, M., El-Daly, S. M., Fabbri, M.
& Calin, G. A. Noncoding RNA therapeutics - challenges and potential
solutions. Nat Rev Drug Discov 20, 629-651,
doi:10.1038/s41573-021-00219-z (2021).
Renganathan, A.
& Felley-Bosco, E. Long Noncoding RNAs in Cancer and Therapeutic Potential. Adv Exp Med Biol 1008, 199-222, doi:10.1007/978-981-10-5203-3_7 (2017).
Pichler, M. &
Calin, G. A. MicroRNAs in cancer: from developmental genes in worms to their
clinical application in patients. British
Journal of Cancer 113, 569-573,
doi:10.1038/bjc.2015.253 (2015).
Kristensen, L. S.,
Hansen, T. B., Venø, M. T. & Kjems, J. Circular RNAs in cancer:
opportunities and challenges in the field. Oncogene 37, 555-565,
doi:10.1038/onc.2017.361 (2018).
Yang, J. et al. CRISPR/Cas9-mediated noncoding
RNA editing in human cancers. RNA Biol 15, 35-43,
doi:10.1080/15476286.2017.1391443 (2018).
Dimitri, A., Herbst,
F. & Fraietta, J. A. Engineering the next-generation of CAR T-cells with
CRISPR-Cas9 gene editing. Molecular
Cancer 21, 78,
doi:10.1186/s12943-022-01559-z (2022).
Hsu, C. L. et al. LncRNA SNHG1 regulates
neuroblastoma cell fate via interactions with HDAC1/2. Cell Death Dis 13, 809,
doi:10.1038/s41419-022-05256-z (2022).
Li, X. et al. microRNA-137 promotes apoptosis
in ovarian cancer cells via the regulation of XIAP. Br J Cancer 116, 66-76,
doi:10.1038/bjc.2016.379 (2017).
Zhao, Z. et al. CircSOD2 induced epigenetic
alteration drives hepatocellular carcinoma progression through activating
JAK2/STAT3 signaling pathway. J Exp Clin
Cancer Res 39, 259,
doi:10.1186/s13046-020-01769-7 (2020).
 Subscribe Us
Subscribe Us Gene Editing Services
Gene Editing Services
 EZ-editor™
EZ-editor™ Red Cotton Gene knockout Project
Red Cotton Gene knockout Project