CRISPR gene editing MC-3T3 cell line
——A useful tool of bone related research
MC-3T3 is a model osteogenic cell line derived from newborn mouse calvaria in 1981. This osteogenic cell line retains its capacity to differentiate into osteoblasts and mineralize in vitro, which makes it a very useful cellular model system for the study of bone biology. The Osteogenic cell line (MC-3T3) has the capacity to produce collagen and differentiate into osteoblast-like cells, forming calcified tissue in vitro and to their in vivo counterparts. Earlier research directed that bone tissue shows constant remodeling by the osteoblastic bone formation and osteoclastic bone resorption in order to maintain bone volume and homeostasis. Therefore, bone remodeling has been suggested to be regulated by crosstalk between osteoblasts, osteoclasts, and osteocytes. MC-3T3 cells have also become important in vitro model for this kind of bone-related research.
Application:
MC-3T3 cells retain the capability to differentiate into osteoblasts. CRISPR/Cas9 based gene editing in MC-3T3 cells can accelerate bone research. MC-3T3 has been widely used in the following types of research:
1. Collagen type 1 biosynthesis, the most important and abundant organic component of the extracellular bone matrix that provides bone strength and flexibility.
2. WNT signaling is an important regulator of bone formation via regulation of osteoblast proliferation and differentiation and matrix mineralization.
3. TGFβ signaling, abnormal bone remodeling, and increased bone turnover.
4. Osteogenic cell differentiation, an osteoblast-specific transcription factor that regulates osteoblast differentiation. Loss of function results in reduced osteoblast differentiation and bone formation.
CRISPR-U™ gene editing in MC-3T3 cells
Genome editing and the creation of cellular models can advance research programs in the area of functional genomics, signaling pathways, metabolism, cell death, drug discovery, drug response, and cancer research, etc. Currently, the Osteogenic cell model (MC-3T3) broadly used for cell differentiation, the effect of paracrine factors for bone formation, or resorption study. Recent findings directed that mutant proteins affecting bone tissue homeostasis and novel pathways are involved in skeletal development and maintenance. To accelerate bone biology research, Ubigene developed CRISPR-U™ for gene manipulation of eukaryotic cells. Thereby, possible to achieve genome editing in MC-3T3 cells with the utilization of the CRISPR/Cas9 system. The CRISPR-U™ system used for rapid and precise gene editing in MC-3T3 cells. CRISPR-generated cell models have enabled mechanism explorations of various skeletal diseases, which are mainly related to skeletal development, resorption, osteosarcoma, and metabolism processes. Based on the elucidation of pathogenic mechanisms, CRISPR-generated cell models could be coupled with drug screening and other therapeutic studies. Ubigene can customize the gene-editing in eukaryotic cells as well as can generate various genes modification in animal models.
New innovation!

Case study:
CRISPR mediated knockout MC-3T3 helps to discover a novel approach to potentially prevent or reverse osteoporosis
Osteoporosis is an enormous, and growing, public health problem. Characterized by low bone mass and defects in bone microarchitecture, osteoporosis leads to increased susceptibility to fracture. Proper maintenance of skeletal health throughout life relies on the coordinate and balanced actions of bone-resorbing osteoclasts and bone-forming osteoblasts, which results in net bone balance. So, there is a clinical need to identify new molecular targets for the treatment of osteoporosis, particularly those that simultaneously inhibits bone resorption while stimulating bone formation. The retinoic acid receptor-related orphan receptor beta (Rorβ) is a novel negative regulator of osteoblastic differentiation and that Rorβ expression is highly elevated in bone marrow-derived osteoprogenitor cells isolated from old, osteoporotic mice, suggesting a potential role for Rorβ in mediating age-related bone loss. Overexpression studies in the pre-osteoblastic mouse cell line, MC-3T3, demonstrated significant regulation of known osteogenic pathways supporting a key role of Rorβ for regulating osteogenesis. The researcher applied CRISPR/Cas9 gene editing to demonstrate that loss of Rorβ in osteoblasts enhances Wnt signaling, specifically through increased recruitment of β-catenin to T-cell factor/lymphoid enhancer factor (Tcf/Lef) DNA binding sites in the promoters of the Wnt target genes Tcf7 and Opg. Thus, resulting in increased osteogenic gene expression and suppressed osteoclast formation through increased osteoprotegerin (OPG) secretion in Rorβ-deficient MC-3T3 cells.
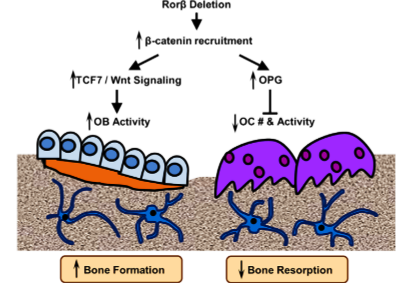
Model of Rorβ action in bone
CRISPR mediated Rorβ mutant generation and gene expression analysis:
The Rorβ gene has two isoforms (Rorβ1 and Rorβ2) and isoform Rorβ2 is not detectable. So, isoform Rorβ1 stands for the Rorβ gene. The ATG start codon of Rorβ1 lies immediately at the 3' end of exon 1; therefore, targeted exon 2 for gene editing. 3 gRNAs were designed and after testing editing efficiency, selected one gRNA to delete Rorβ in MC-3T3 cells. A control cell line “MC3T3-Cont” produced by using a nonspecific gRNA. Cloning and sequence analysis of the MC3T3-DRorβ cell line revealed a 1-basepair (bp) and 4-bp deletion in the mouse Rorβ alleles, which leads to frameshift mutations that ultimately resulted in nonfunctional alleles. To analyses, the effects of Rorβ deletion on osteoblastic gene expression, collect RNA samples at various time points from the MC3T3-Cont and MC3T3-DRorβ cells grown in an osteogenic medium. qPCR analyses revealed that significantly increased expression of bone marker genes in MC3T3-DRorβ cells.
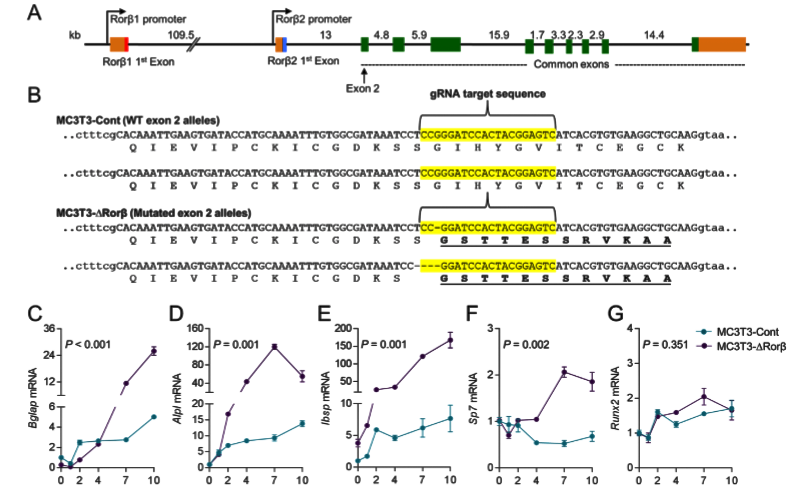
Fig 1: CRISPR/Cas9 deletion of Rorβ in MC3T3-E1 pre-osteoblastic cells. (A) The intron/exon structure of the mouse Rorβ gene is shown. (B) Exon 2 (in uppercase) of the mouse Rorβ gene was mutated using CRISPR/Cas9 gene editing methodology. The guide RNA (gRNA) sequence used to target the allele is highlighted in yellow. (C-G) both cell models were treated with osteogenic medium at confluence and qPCR for the donated bone markers were performed on samples were collected at days 0 to10 (n=6).
Rorβ-deficient osteoblasts display enhanced Wnt signaling through β-catenin–dependent mechanisms:
The mutant MC3T3-DRorβ cells were used to identify altered canonical cellular pathways. They observed a significant alteration in the Wnt/β-catenin pathway and expression of several well-known regulators of this pathway that were modulated in MC3T3-DRorβ cells. IPA-analysis showed significant enrichment in T-cell factor/lymphoid enhancer factor (Tcf/Lef) DNA binding sites. These findings indicate that loss of Rorβ in osteoblastic cells leads to alterations in the Wnt/β-catenin pathway. Two well-known Wnt target genes, Opg and Tcf7 were verified by RNAseq and observed that this gene expression was significantly upregulated in Rorβ-deficient cells throughout the time course. The role of the Wnt pathway in Opg and Tcf7 upregulation in Rorβ-deficient osteoblasts, a series of inhibitor studies using either DKK1 or JW55 and they found that the Wnt pathway is altered by the loss of Rorβ.
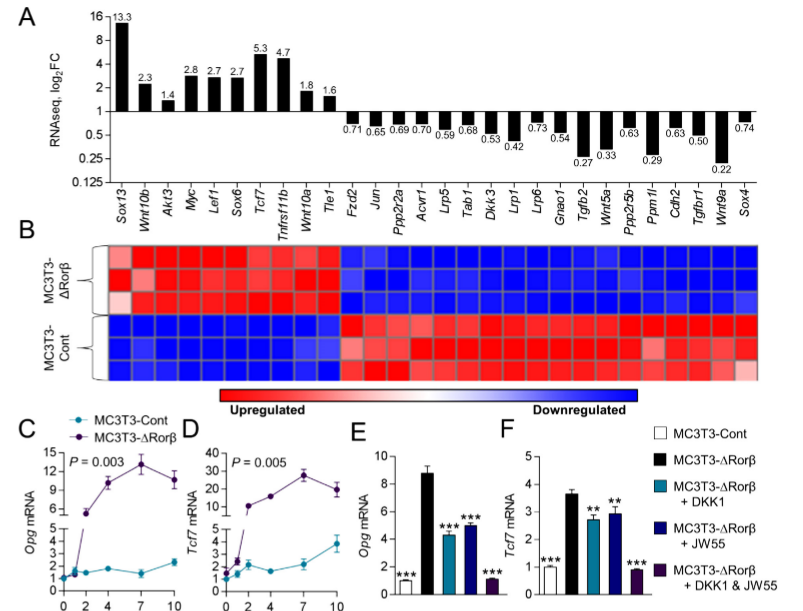
Fig 2: Rorβ regulates the Wnt/βcatenin pathway. (A, B) Wnt/βcatenin regulated gene identified by RNAseq and IPA analysis are significantly regulated in MC3T3 DRor β. (C, D) The time course from fig.1was re-queried and analyzed for Opg and Tcf expression. (E, F) The MC3T3-Cont and MC3T3-D Rorβ cell lines were treated with osteogenic medium at confluence and then supplemented with either Dkk1 (200ng/mL), JW55 (10mM), or both; cells were harvested 48 hours later and qPCR was performed for Opg and Tcf7 (n=6).
Rorβ-deficient cells suppress osteoclastogenesis by OPG production:
ChIP assays on the MC3T3-Cont and MC3T3-DRorβ cells were performed to understand the mechanism of β-catenin in the regulation of the Opg and Tcf7 genes. The researcher found that β-catenin and RNA polymerase (RNAP) recruitment was increased at Tcf/Lef DNA binding sites located within the promoters of the Opg and Tcf7 genes in the Rorβ-deficient cells. They verify that Rorβ inhibits the transcriptional activity of a constitutively active form of β-catenin (ca-βcat) on the TOP-FLASH Wnt-reporter construct. So, they conclude that Rorβ inhibits Wnt activity through the inhibition of β-catenin recruitment to Tcf/Lef binding sites in the promoters of Wnt responsive genes in osteoblasts. They assessed the implications of increased OPG on osteoclastogenesis. They found that Rorβ-deficient cells are capable of suppressing osteoclastogenesis through increased OPG production and secretion.
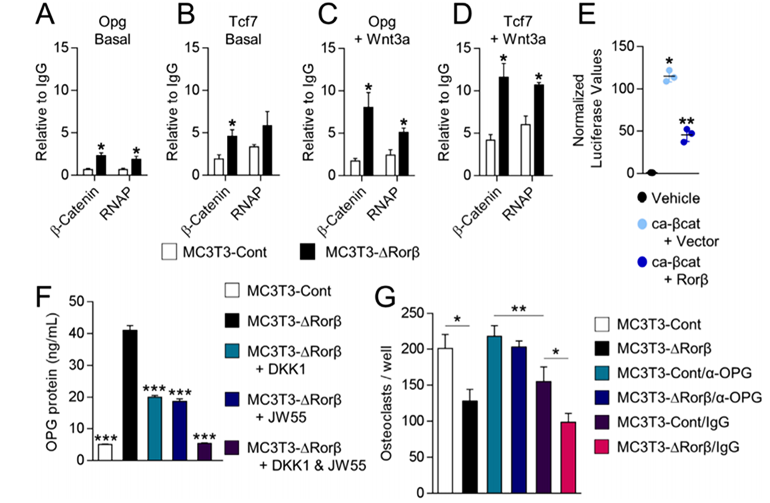
Fig 3: Rorβ-deletion enhances β-catenin recruitment to the promoters of Wnt-responsive genes. (A–D) ChIP assays were performed on the MC3T3-Cont and MC3T3-DRorβ cell lines following 6-hour treatment with 10 ng/mL Wnt3a, or untreated (Basal). IPs were performed for β-catenin and RNAP and QPCR specific for the Tcf/Lef DNA binding site in the promoters of the Opg and Tcf7 gene. The data are presented relative to an IgG negative control IP. (E) Ca-βcat and TOP-FLASH were co-transfected –/+ Rorβ and luciferase activity was measured 48 hours later (n = 6). (F) CM from the treated cells was collected and OPG protein levels were determined by ELISA. (G) Bone marrow-derived osteoclast precursors were incubated with osteoclastogenic-supportive medium supplemented with 1% of the culture media.
Summary of the current understanding of the osteoprotective effects of the deletion of Rorβ, which triggers a multifaceted response in the bone microenvironment. The deletion of Rorβ enhances bone formation through increased Tcf7 and Wnt signaling responses. Concurrently, Rorβ deletion affects bone resorption through increased production and secretion of OPG, which reduces osteoclast numbers and osteoclast activity. Collectively, the increase in bone formation and a decrease in bone resorption results in the prevention of bone loss. The inhibition of Rorβ may represent a novel approach to prevent or treat osteoporosis.
CRISPR-U™ efficiently modify genes in MC-3T3 cell line:
CRISPR-U ™ is an exclusive technology independently developed by Ubigene Bioscience for gene editing cell lines. By optimizing gene editing vectors and processes, the efficiency in gene-cutting and recombination of CRISPR-U ™ is 10 times higher than the conventional CRISPR/Cas9 technology. We are capable of customizing genetically-modified MC-3T3 cell line as you desire and satisfying various needs in gene editing. We offer our clients for knockout, knockin and point mutation based modified MC-3T3 model cells for research.
References
1. Osteoprotection Through the Deletion of the Transcription Factor Rorb in Mice. Journal of Bone and Mineral Research, 33(4) 2018, 720–731.
 Subscribe Us
Subscribe Us Gene Editing Services
Gene Editing Services
 EZ-editor™
EZ-editor™ Red Cotton Gene knockout Project
Red Cotton Gene knockout Project