[March Colorectal Cancer Awareness Month] CRISPR helps to discover better targets of CRC

Colorectal cancer (CRC) is a heterogeneous intestinal epithelial disease characterized by gene mutation accumulation and immune response disorder. It is the third most common cancer and the second highest cancer mortality in the world. This disease poses a serious threat to human health. Therefore, studying the pathogenesis and treatment of CRC to facilitate the clinical diagnosis and treatment of CRC has become an urgent demand of the world.
In recent years, with the rapid development of molecular biology and genetic engineering technology, the mystery of human diseases, including CRC, is being gradually unveiled. Among them, the gene engineering technology with CRISPR/Cas9 is widely favored by researchers, and has become the key technology to discover the key factors involved in CRC regulation and develop potential therapeutic targets. We’ve selected the recent CRC researches with gene editing, hope these studies could inspire you.
Case study 1: Genome-wide CRISPR-cas9 knockout screening identifies GRB7 as a driver for MEK inhibitor resistance in KRAS mutant colon cancer. Oncogene, 2022Targeting KRAS pathway is a promising and challenging treatment for colorectal cancer. Although MEK inhibitor (MEKi) has shown efficacy in BRAF mutant melanoma, drug tolerance due to intracellular compensatory signal transduction in CRC cells limits its clinical application in CRC. Therefore, it is significant to identify the key genes related to MEKi resistance and provide combination therapy to overcome MEKi resistance.
This study designed a combined strategy, including genome-wide CRISPR/Cas9 knockout library screening and transcriptome analysis of KRAS mutant cell line (HCT116). The results showed that EGFR, MET, FAK, STAT3 and AKT were identified as candidate genes related to MEKi resistance, and they were significantly enriched in GRB7 mediated RTK pathway.

To verify the key role of GRB7 in MAPKi resistance, the researchers evaluated the proliferation of KRAS mutant CRC cells in the presence of MAPKi after shRNA knockdown of GRB7. The results showed that after GRB7 knockdown, the proliferation of CRC cell lines (HCT116, SW480, LS174T) decreased significantly in the presence of MAPKi, and the apoptosis accelerated. In addition, the acquisition of resistance in CRC model cell lines was studied by overexpression of GRB7. It was found that after GRB7 gene overexpression, the clonogenic ability of CRC cells was significantly improved, and the proportion of apoptosis decreased. This series of results suggest that GRB7 regulates MAPKi resistance in KRAS mutant CRC cells.
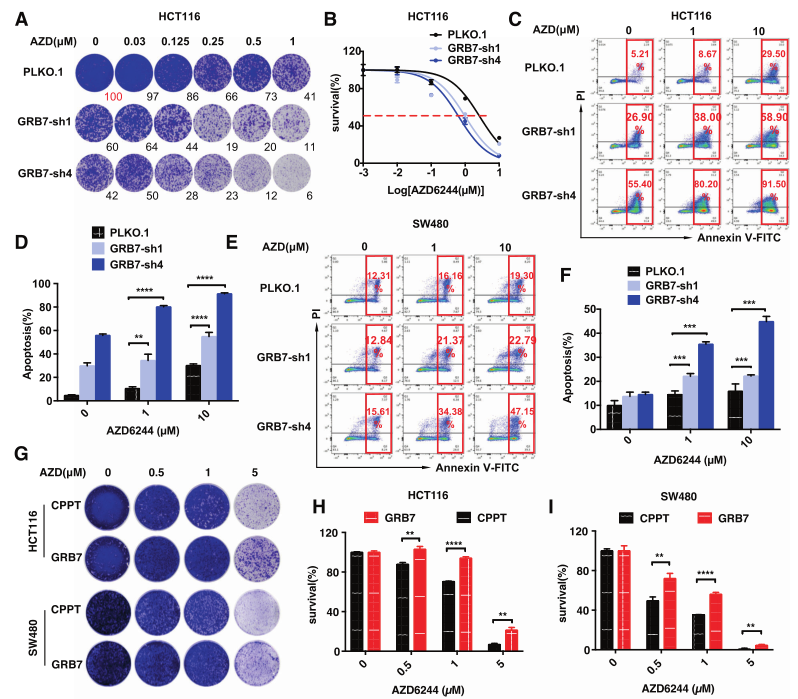
Further, the researchers determined that PLK1 was the main interacting kinase of GRB7 based on immunocoprecipitation and mass spectrometry (IP-MS), and confirmed that the combination of PLK1 and MEK inhibitors synergistically inhibited CRC cell proliferation and induced cell apoptosis in vitro and in vivo in 2D and 3D CRC models.
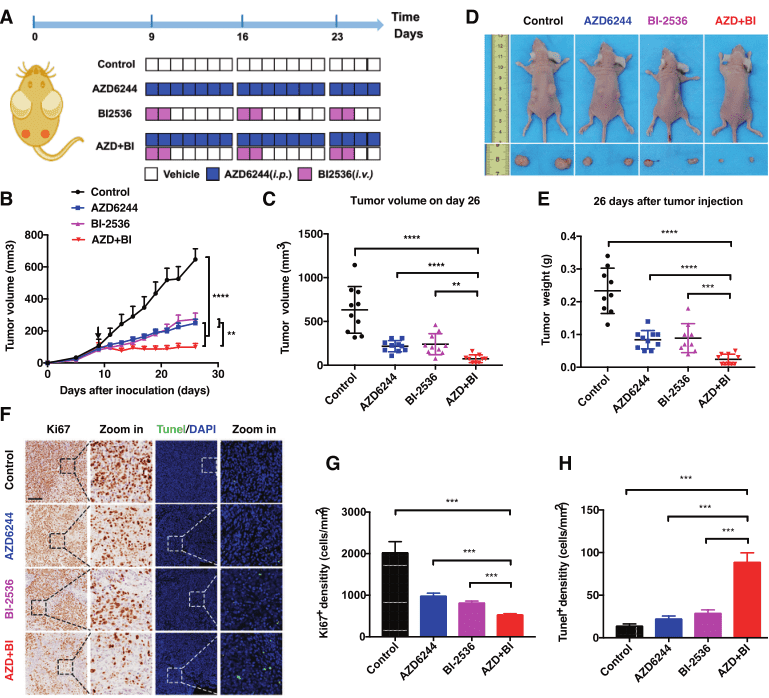
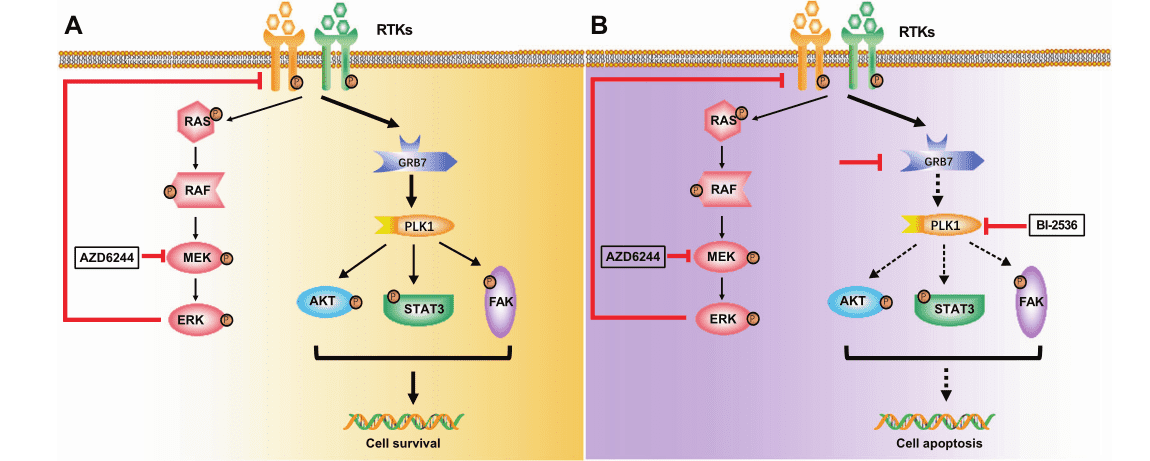
In conclusion, this study identified GRB7-PLK1 as the core axis mediating RTK, leading to MEKi drug tolerance. Therefore, PLK1 combined with MEKi is a potential target for the clinical treatment of CRC patients with KRAS mutation.
Ubigene provides customized cell services for GRB7 knockdown HCT116, SW480 and LS174T, which are used in the above research. Starting from $1780, unlock the knockdown guaranteed custom stable cell line service now!
In order to speed up cancer diagnosis and treatment research, Ubigene screened nearly 1000 high-frequency mutation loci, including mutations in KRAS and EGFR. It only takes $6480 to get a point mutant cell line!
Click here to get the list of these mutations>>Your target is not in the list? Check out Ubigene’s new Red cottonTM CRISPR point mutation designer, get the optimal strategy in 1 min,
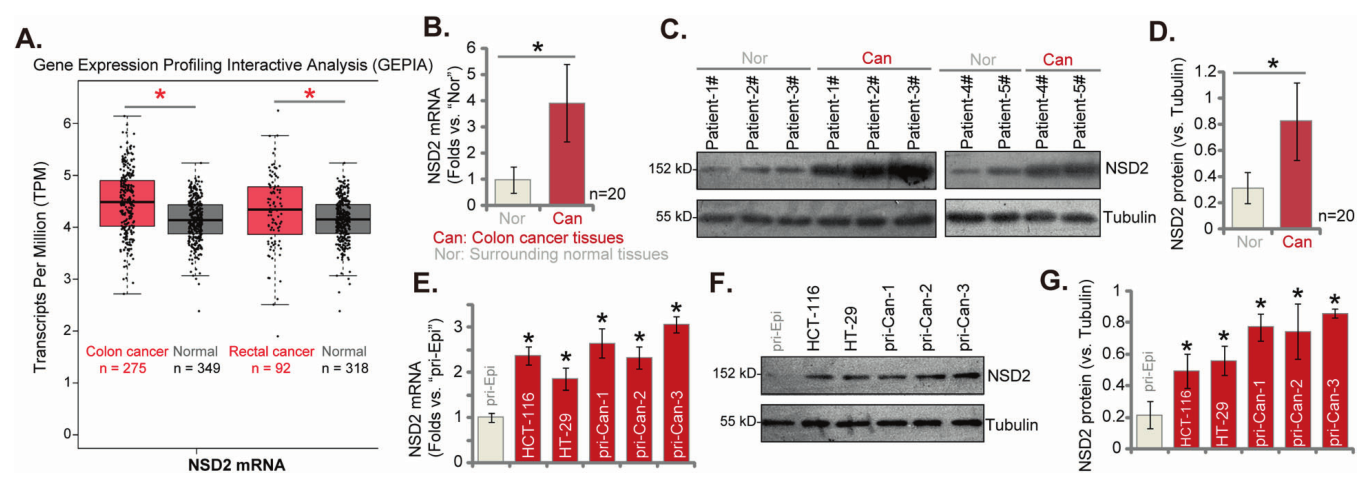
click here to access>>Case study 2: Identification of histone methyltransferase NSD2 as an important oncogenic gene in colorectal cancer. Cell Death & Disease, 2022Histone 3 (H3) methylation may lead to the transcriptional activation of many oncogenes, which is closely related to the development and process of CRC. Nuclear receptor SET domain protein 2 (NSD2) is a key histone methyltransferase that catalyzes histone H3 lysine 36 dimethyl (H3K36ME2). Studies have shown that NSD2 is overexpressed in several solid tumors. However, the expression, potential function and molecular mechanism of NSD2 in CRC have not been reported.
In this study, bioinformatics results from the GEPIA (Gene Expression Profiling Interactive Analysis) showed that NSD2 mRNA expression was elevated in colon and rectal cancer. In addition, the expression levels of NSD2 mRNA and protein in local colon cancer tissues were significantly higher than those in the surrounding normal tissues.
In shRNA mediated silencing or CRISPR/Cas9 mediated NSD2 knockout primary human colon cancer cells and NSD2 knockout CRC cell lines, cell proliferation, cell cycle progression, migration and invasion were significantly inhibited.
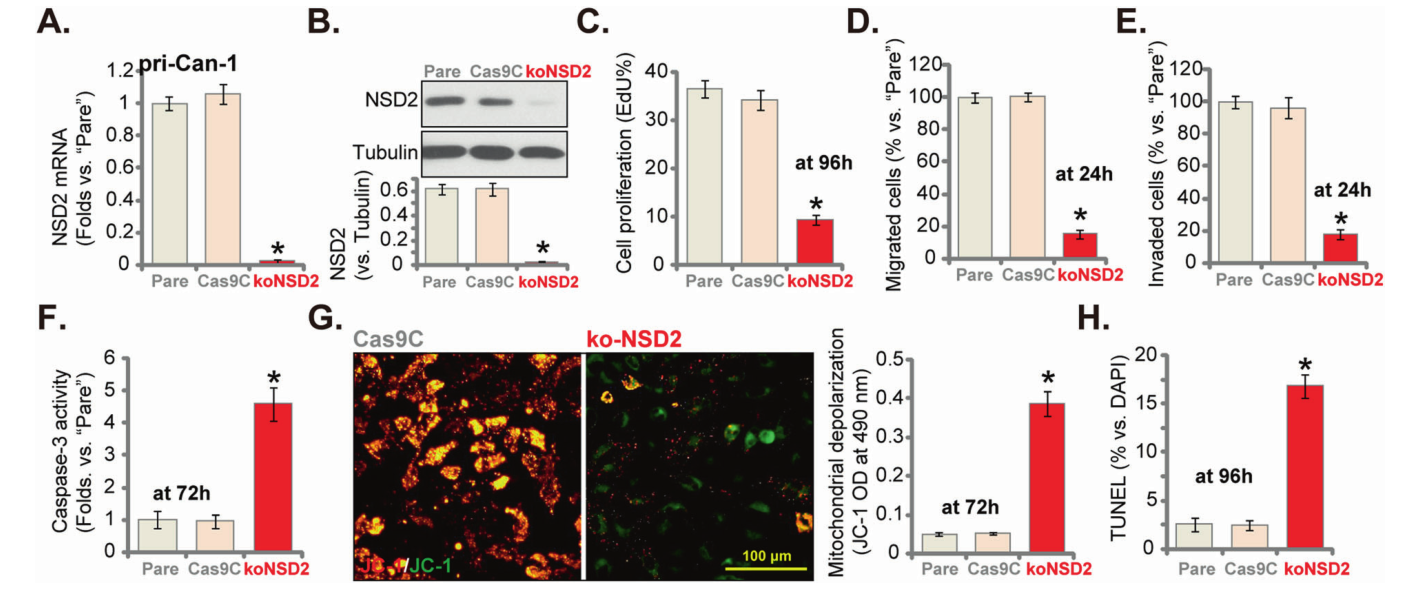
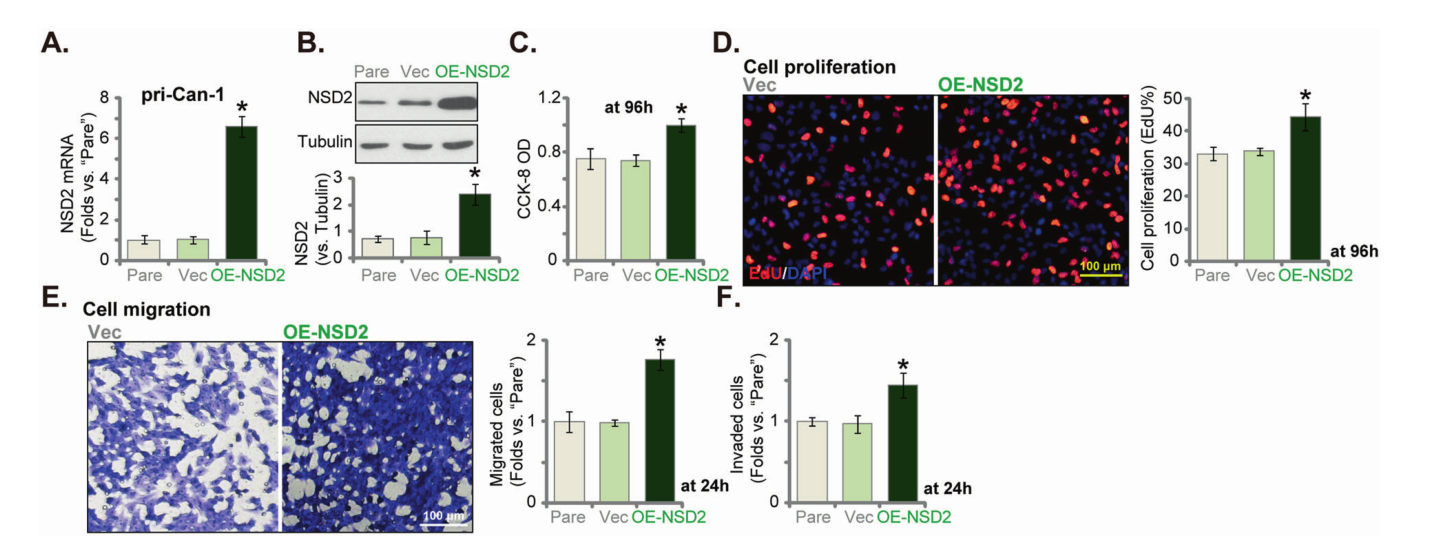
Conversely, in the primary human colon cancer cell line (pri-Can-1) with NSD2 overexpression, the ability of cell proliferation, invasion and migration increased significantly.
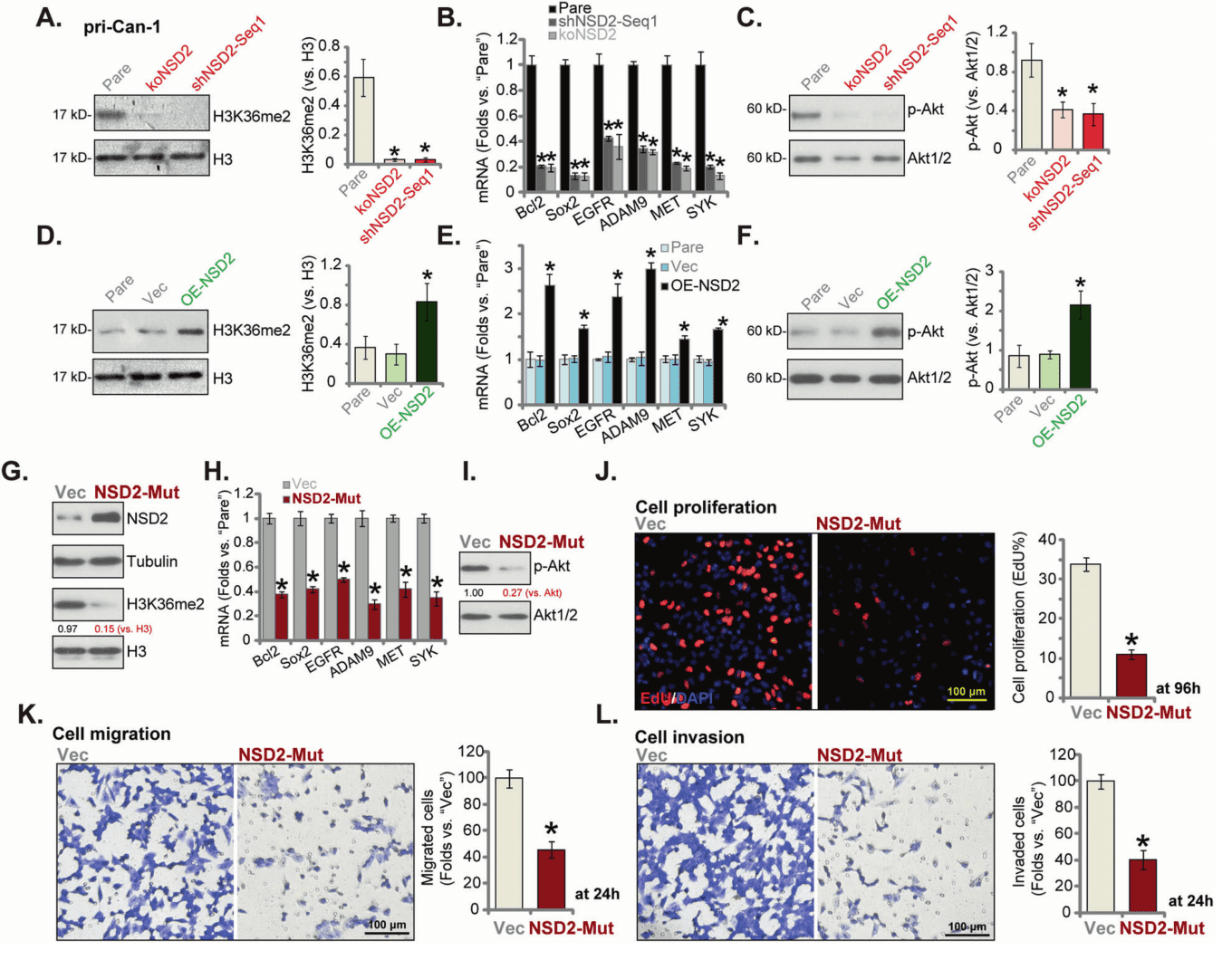
In further research, the researchers found that the level of H3K36me2 and the expression of NSD2 related oncogenes ADAM9, EGFR, Sox2, Bcl-2, SYK and MET were largely inhibited in NSD2 silenced or knockout primary human colon cancer cells. However, their levels increased with NSD2 overexpression.
In vivo intraperitoneal injection of AAV carried NSD2 shRNA can inhibit the growth of primary colon cancer cells in nude mice to a great extent, and quantitative analysis found that the level of H3K36me2 and the expression of oncogene decreased significantly.
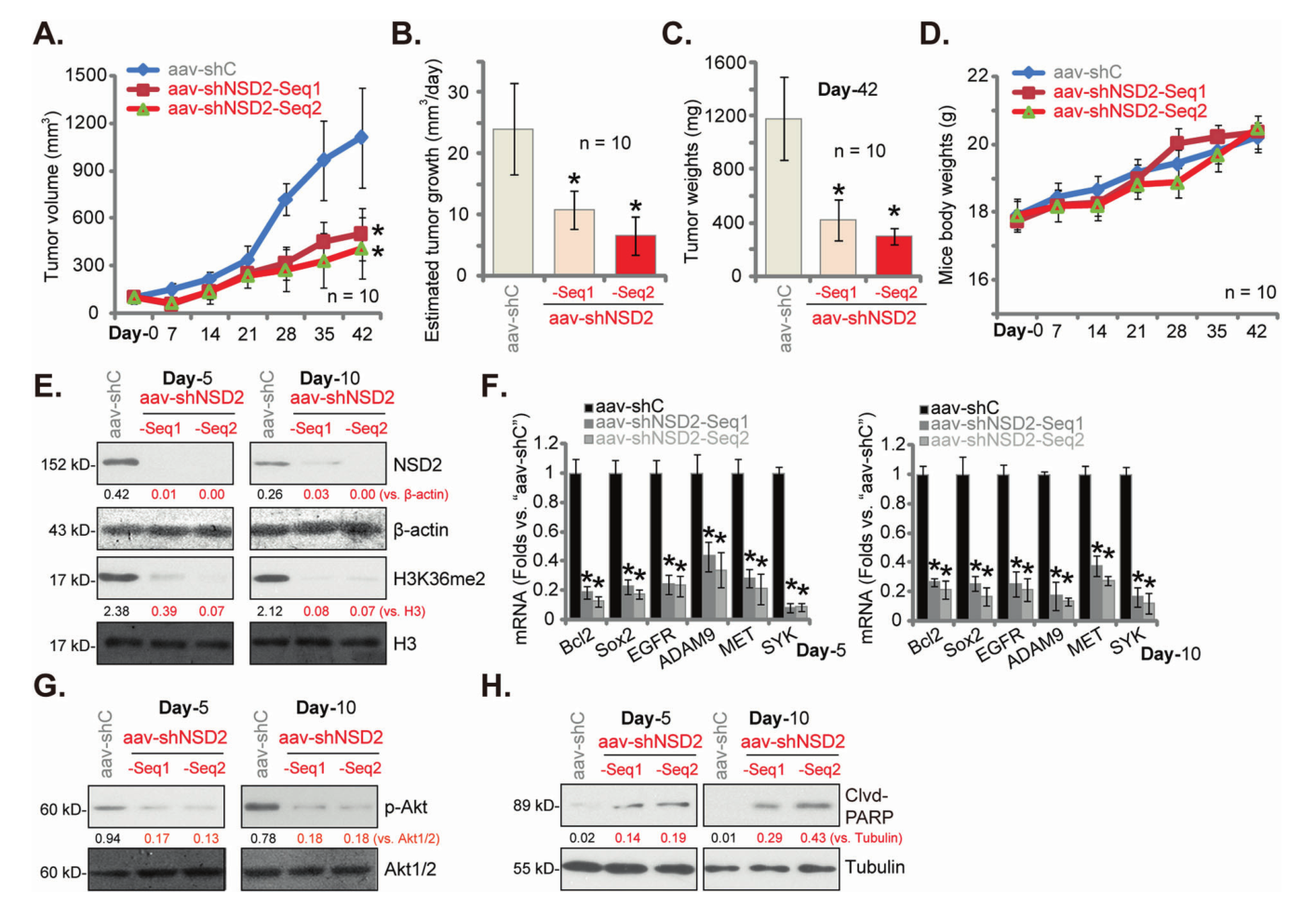
Therefore, NSD2 is a new important oncogene required for CRC growth in vitro and in vivo. Targeting NSD2 may be a potential strategy for CRC treatment.
Silencing or overexpression of genes has become the must-have method to study gene function. Ubigene offers custom stable cell line services and AAV with interference/overexpression of the above genes (
click to learn about our back-to-school sales>>). We also provide a variety of plasmid in stock, such as NSD2 gene knockout plasmid mentioned in the study.
Click to search for your desired plasmid >>No time to construct KO cell lines by yourself? Hand it over to Ubigene! For only $2021, KO cell line can be delivered in 8 weeks.
Ubigene is an enterprise focusing on gene editing. We provide high-quality gene editing cell lines, stable cell lines, virus packaging and other related services worldwide. Ubigene has successfully edited thousands of genes in about 200 cell lines, and successfully constructed a Knockout Cell Line Bank with nearly 3000 genes. Feel free to contact our scientists to discuss your projects.
 Subscribe Us
Subscribe Us Gene Editing Services
Gene Editing Services
 EZ-editor™
EZ-editor™ Red Cotton Gene knockout Project
Red Cotton Gene knockout Project

















